The focus is increasingly moving towards vaccine roll-out as the world faces the real possibility of the second year of social and economic restrictions. The vaccine may help slow down the viral spread and reduce the mortality and morbidity associated with this infection.
Several vaccines are at or near the finishing post already, mostly directed against the viral spike antigen. However, there is a worrying increase in the emergence of escape mutations that allow the virus to evade these antibodies' neutralizing effects. A new study published in the medRxiv* preprint server indicates a critical cause of the positive selection pressure that underlies this phenomenon.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has several structural proteins, including the key spike glycoprotein. This protein is among the dominant antigens and crucial for viral entry into the host cell. It is the spike receptor-binding domain (RBD) that is bound by neutralizing natural and therapeutic monoclonal antibodies.
Several mutations have been documented in the viral genome over the pandemic's year-long course, of which some have been particularly successful. This includes the spike D614G mutation, which has rapidly risen to the dominant position in every place where it was introduced, displacing the original strain. This mutation confers a definite advantage on the virus in terms of infectivity and transmissibility, accounting for the rapid spread of strains characterized by it.
Deletion mutations have also been recognized in different strains within the same host and across different individuals. The most important is H69/V70, a double deletion found in multiple separate strains in Europe.
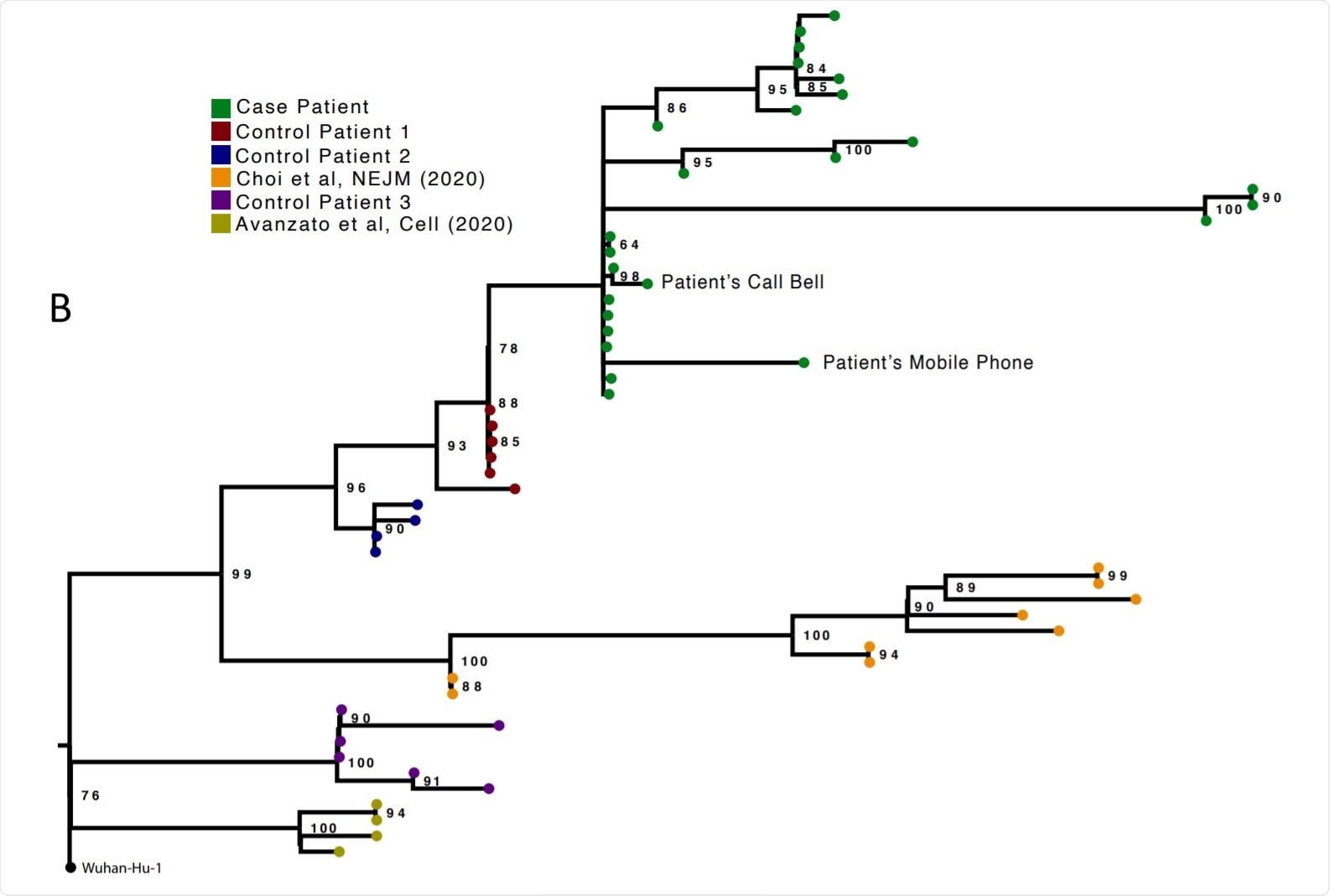
Analysis of 23 Patient derived whole SARS-CoV-2 genome sequences in context of national sequences and other cases of chronic SARS-CoV-2 shedding. Image Credit: https://www.medrxiv.org/content/10.1101/2020.12.05.20241927v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study Details
The current study follows the emergence of this double mutation in combination with the S2 mutation D796H and broad escape from antibody neutralization in a patient who received convalescent plasma therapy. The patient was immunocompromised because of treatment with the B cell depletion agent rituximab.
The study included the measurement of specific anti-SARS-CoV-2 antibodies over time. The initial negative results at day 44 and day 50 following infection were followed by an infusion of three units of convalescent plasma (CP) from three separate donors. These had specific neutralizing antibodies targeting the virus.
The researchers compared genomic analyses from 23 successive respiratory samples, mostly from post-intubation nose and throat samples or endotracheal aspirates. Ct values were between 16 and 24. Samples from the call bell and the telephone in the patient’s room yielded viral isolates from the same population on days 59, 92 and 101, indicating environmental contamination. The infecting strain was of the lineage 20B with the D614G mutation in the spike.
Over 101 days, the virus population showed marked diversity of viral strains, with the mutation rate being estimated at 30 per year. At days 93 and 95, the viral strains were significantly different from the early isolates, though closely related phylogenetically. Rather than being the effect of positive selection pressures, the investigators infer that this reflects rapid mutagenesis within a small subset due to enhancement by genetic drift.
Sequential samples from three other patients who also had viral RNA shedding for four or more weeks were also sequenced, but interestingly, the compositional shift did not occur in these cases, especially concerning the spike protein.
Escape in Association with Therapy
Two courses of remdesivir therapy (by day 54) and two CP infusions (by day 65) failed to change the Ct values up to the first 100 days post-infection. After day 66, the viral population showed a drastic compositional shift.
By day 82, the D796H mutation in S2 became almost the sole allele, along with the S1 double mutation H69/V70. This set of mutations then dropped to a very low frequency by days 86 and 89, while spike mutations Y200H and 163 T240I became predominant. Despite the clear divergence, this was not due to superinfection, as the phylogenetic relationship was observed.
On day 93, the set of D796H along with H69/V70 was detected in less than 10% of the samples in a nose and throat sample. Simultaneously, a group of viruses with spike RBD mutation P330S and S1 NTD mutation W64G had increased to almost 100% abundance at this date. The RBD mutation was present in two distinct samples. This divergent sample resulted from the stochastic development of a hitherto undetected subgroup of viruses in the upper respiratory tract.
After the third course of remdesivir on day 92 and the third CP infusion on day 95, the D796H + H69/V70 strain increased. The researchers consider this to be the effect of positive selection pressure renewed by the third unit of CP. The level of inflammation was hyperintense, but Ct values were low through this period, terminated by the patient's death on day 102. The cause of death was multi-organ failure.
The repeated increase in frequency of the novel viral strain during CP therapy strongly supports the hypothesis that the deletion/mutation combination conferred antibody escape properties.”
Broad Escape Mechanism
The scientists found that the neutralization activity of CP against the wild-type was higher than against the H69/V70 + D796H mutant D614G-carrying spike protein, as tested in a pseudoviral system.
After two CP units, the triple mutant continued to persist in the patient’s samples. Patient serum both before and after the second unit of CP showed only partial neutralization efficacy against this variant. The persistence and re-emergence of this variant after the third unit of CP emphasizes its lack of susceptibility and indicates a broad escape mechanism.
The researchers tested five randomly selected sera from recovered patients to neutralize activity against wild-type and mutant viruses. The mutant strain resisted neutralization by four of the five sera, with non-significant neutralization activity by the fifth. In contrast, the wild-type virus was up to ten times as susceptible as the mutant strain.
What would be the impact of monoclonal antibodies on the mutant virus? To find out, the researchers looked at the neutralization activity of a seven-antibody set to an array of epitope groups. There was no difference in the neutralization achieved with any of the epitope clusters. The researchers concluded that the mechanism of immune escape did not belong to these epitopes.
Mutations Affect Antibody Binding
They then analyzed a published spike structure, annotating the residues related to the escape. They found the double mutation affected a disordered loop at the tip of the NTD, with multiple sugar residues attached. Therefore, the location could easily disrupt antibody binding. Secondly, the D796H mutation is relatively uncommon. However, because it affects an exposed loop in the S2 spike subunit, a part of the spike that is an important epitope could interfere with antibody binding.
In the current case, the change in viral population genes in response to CP was capable of being followed over the long-term because of the prolonged viral shedding due to persistent viral replication and the absence of antiviral effect despite remdesivir and CP infusion. Such patients are prone to developing prolonged infections.
What are the Implications?
Overall, the study shows how the genetic makeup of the virus population shifts with the administration of CP. In response, a viral strain emerged carrying a beneficial mutation, which became less abundant, before again rising when exposed to the next unit of CP – a feature of positive selection.
The study's implications are potentially alarming, in that it suggests the repeated emergence of a resistant SARS-CoV-2 variant, which has been showing a rise in frequency in Europe. This patient's death may not have been due to viral proliferation following the emergence of this escape mutation. However, the scientists say, “Given that both vaccines and therapeutics are aimed at Spike, our study raises the possibility of virus evasion, particularly in immune-suppressed individuals where prolonged viral replication occurs.”
Our data suggest caution in use of CP in patients with immune suppression of both T cell and B cell arms. In such cases, the antibodies administered have little support from cytotoxic T cells, thereby reducing chances of clearance and raising the potential for escape mutations.”
Secondly, in such cases, the patient should be in a single room with negative pressure and high air-change frequency because of the high chances of environmental contamination, as demonstrated in this case.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources