The Oxford Vaccine Group is in high demand at the moment as they make huge progress in their goal to finalize a vaccine for COVID-19. However, elsewhere in the group, other work is going on that also has wide implications for COVID-19 and the future of vaccine development.
Adriana Tomic, co-developer of SIMON, an open-source platform for applying machine learning to biomedical data, has given News-Medical insight into this groundbreaking research on flu vaccine responses.
 Image Credit: People Image Studio / Shutterstock.com
Image Credit: People Image Studio / Shutterstock.com
Why is the flu vaccine such an important problem to solve?
Millions of people are infected with the influenza virus, commonly known as flu, every year, and whilst for most, suffering is short-term, what many do not realize is that for some people, it can be fatal. It is estimated that every year flu kills up to 0.5 million people worldwide, and especially vulnerable are elderly and young children.
Due to a high mutational rate, new flu strains appear regularly, making the development of a vaccine particularly challenging. Each year, we have to develop flu vaccines to provide protection against the circulating viral strains otherwise there is a risk of a global pandemic. Therefore, to prevent future pandemics, it is of critical importance to understand how seasonal vaccines provide protection against flu.
Using machine learning to study human vaccine responses
Tomic describes SIMON, or Sequential Iterative Modeling OverNight, as an “open-source software for the application of machine learning to biomedical data”.
The Oxford Vaccine Group set about with the goal of improving the vaccine’s effectiveness by determining who was likely to respond positively to it.
The major aims of our project are to understand why some individuals who receive the vaccine are protected against flu while others get infected and to predict if an individual will respond appropriately to the vaccine.”
Adriana Tomic, Ph.D., Oxford Vaccine Group
Using SIMON to understand these challenges involved a lot of time-consuming work, processing and standardizing all the clinical data before the analysis. However, the results have proved it was worth it.
SIMON was able to identify immune cell subsets that were present in those who would respond well to the vaccine, described in one of the most-read publications of 2019 and 2020 as rated by the American Association of Immunologists. These results came from FluPRINT, a project in collaboration with Stanford University that analyzed five flu clinical studies from 2007-2015.
How could these findings improve flu vaccines?
SIMON can be used to predict if the individual can respond appropriately to a vaccine based on a set of immune system parameters. By using SIMON, we identified subsets of immune cells not previously described to provide protection against the virus.
These results are important for the development of the next generation of vaccines and have the capacity to fundamentally change vaccinology by application of computational discovery algorithms to speed up the discovery of the biomedical patterns.”
Adriana Tomic, Ph.D., Oxford Vaccine Group
The theory behind this discovery for improving the flu vaccine is that the cell types that are responding well in some individuals could be ameliorated in those who are not responding well. This offers huge potential in improving the effectiveness of the flu vaccine.
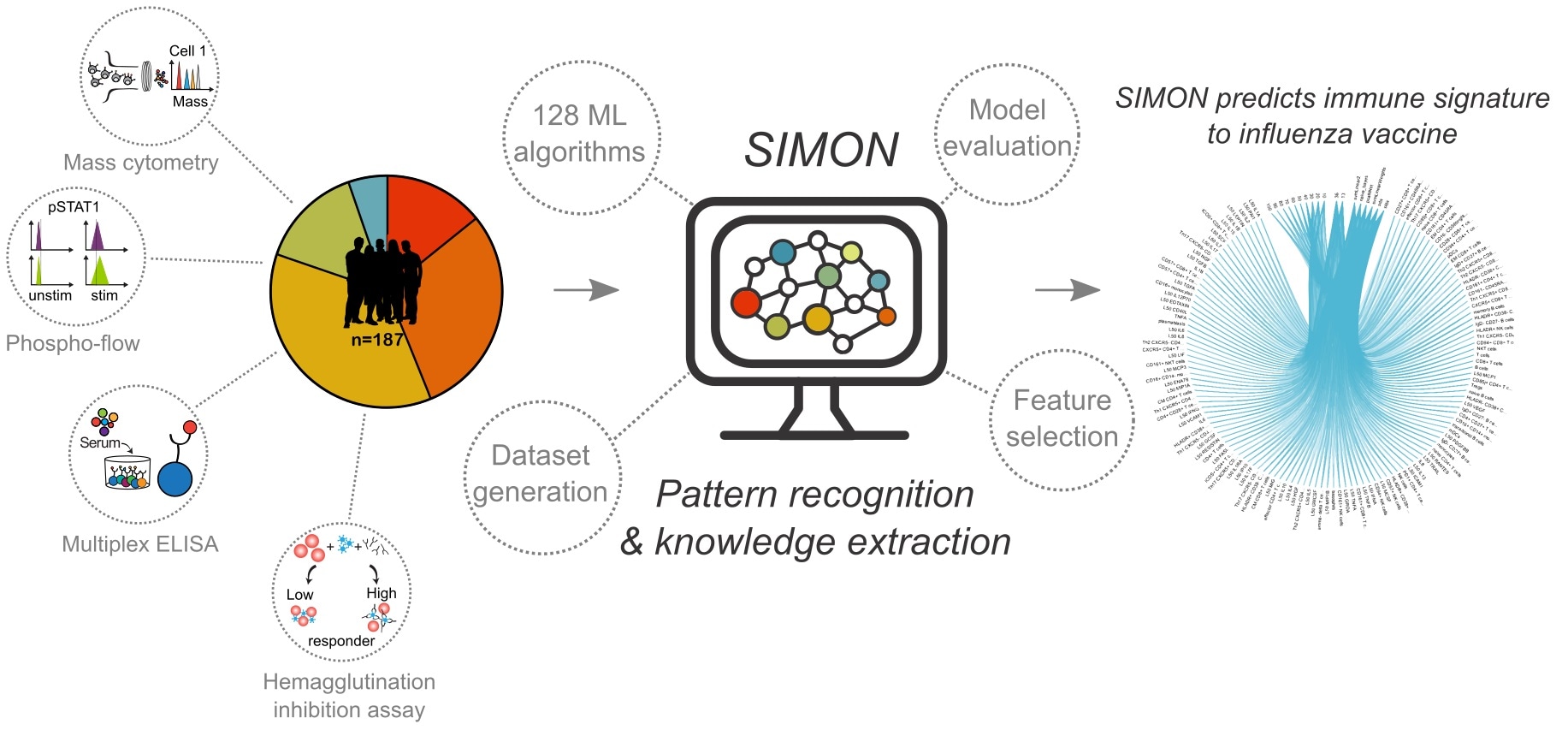 SIMON - the open-source platform for applying machine learning to biomedical data. Image Credit: Adriana Tomic
SIMON - the open-source platform for applying machine learning to biomedical data. Image Credit: Adriana Tomic
Making SIMON accessible
SIMON was downloaded more than 3,000 times and it is widely used by immunology and vaccinology specialists across the University of Oxford, Stanford University, including also researchers at the Walter and Eliza Hall Institute for Medical Research in Melbourne. The software is open-source and free to download, as Tomic and the other developers believe it should be accessible to all in order to have the best impact.
What is next for SIMON?
Currently, SIMON is being used to investigate the flu vaccine which is given to children and understand whether re-immunization reduces protection. If this is proved true, it will greatly affect when flu vaccines should be given.
Vaccines for other diseases are also being examined. The Oxford Vaccine Group is also looking into meningococcal vaccine given to infants and vaccines against Salmonella typhi.
Why is this so important in relation to COVID-19?
Adriana Tomic is part of the team developing a potential vaccine for COVID-19, which is well on its way to approval.
Our work with SIMON and systems immunology approach might be crucial for understanding SARS-CoV2 and the development of the vaccine to stop COVID-19 pandemics.”
Adriana Tomic, Ph.D., Oxford Vaccine Group
SIMON could be a key factor in supporting this vaccine by applying machine learning to the immunological data being gathered around the world. It may help to develop more targeted and efficient therapies for COVID-19.
“Another important aspect is an open-access FluPRINT database which we generated from multiple clinical trials on flu vaccines performed at the Stanford University,” Tomic writes. “Generating similar datasets from ongoing clinical trials on SARS-CoV2 will be critical for understanding this virus and generating an efficient vaccine.”
Beyond the current vaccine, this research could have larger implications, as it may be used in vaccine development for all sorts of diseases. This is a promising prospect, given the likelihood of increasing numbers of new diseases in our future and hence the potential for pandemics to become more common.
To view the SIMON website, click here (genular.org).
Further Reading
The entire FluPRINT project is described in detail on the following website: www.fluprint.com
The publication on SIMON is available at:
https://www.jimmunol.org/content/jimmunol/early/2019/06/13/jimmunol.1900033.full.pdf
Access Adriana Tomic’s review of how the dataset was generated here: https://researchdata.springernature.com/channels/1455-behind-the-paper/posts/53950-unifying-clinical-data-to-reveal-influenza-imprint-on-immune-system.
The publication for the open-access FluPRINT database is available at: https://www.nature.com/articles/s41597-019-0213-4.
About Dr. Adriana Tomic

Adriana Tomic, Ph.D. is a Marie Curie Fellow working at the Oxford Vaccine Group where she leads a systems immunology research aimed at applying machine learning to understand how vaccines work. During her postdoctoral training with Dr. Mark M. Davis at Stanford University she developed a computational tool for the application of machine learning to clinical datasets.
As a Ph.D. student at the Hannover Medical School, Germany, Adriana generated a novel cytomegalovirus vaccine and for this work, she received the award for the best Ph.D. work.