The spread of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causing the coronavirus disease 2019 (COVID-19) pandemic, has led to hundreds of thousands of deaths.
Over the past year, much research has focused on the factors that drive the spread and mutation of the viral lineages in each region. A recent preprint that appeared on the bioRxiv* server applies a new approach to conventional birth-death lineage modeling. Focusing on strains currently circulating in the USA, the study shows the contribution of different factors to viral fitness.
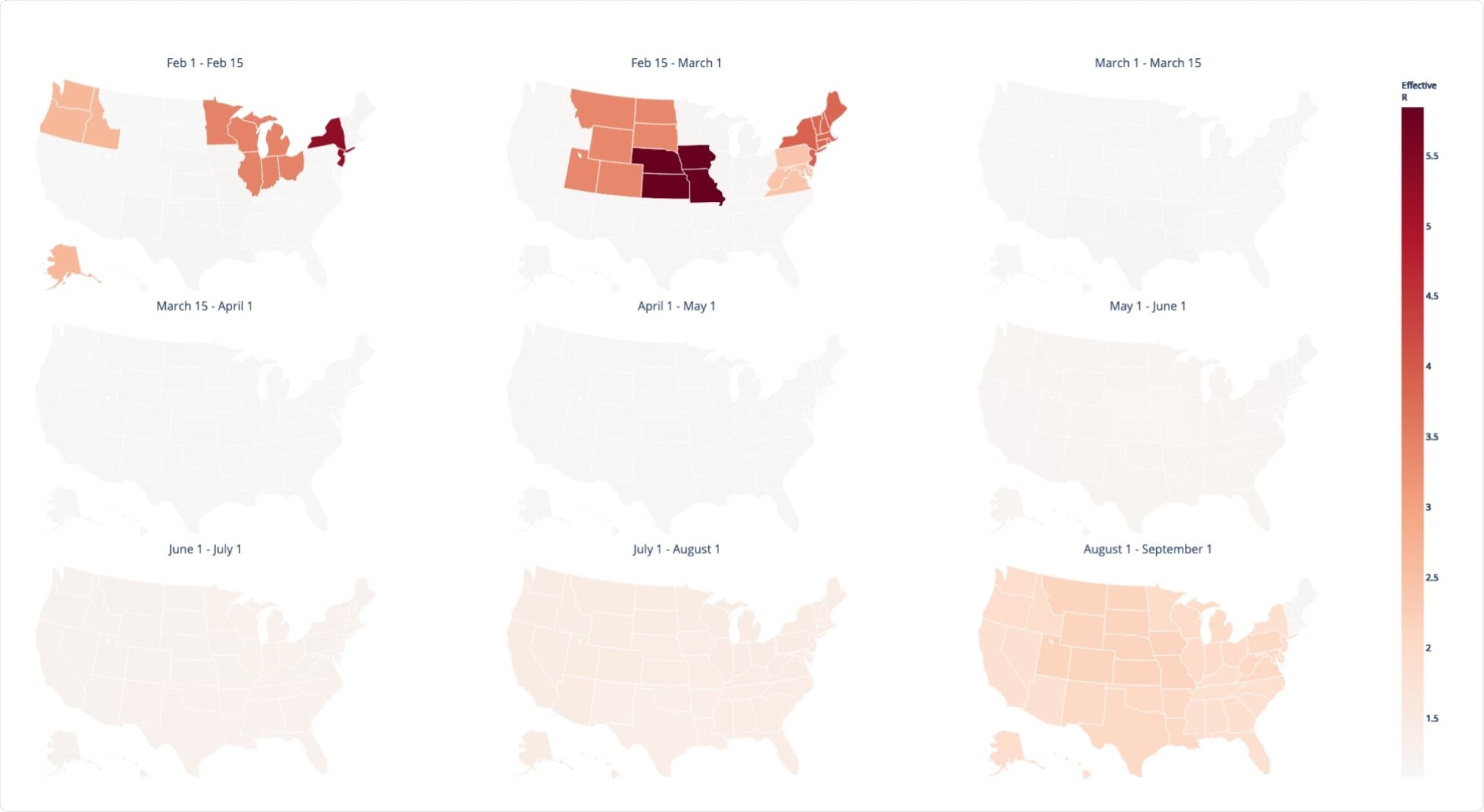
Background spatiotemporal heterogeneity in the effective reproductive number Re of SARS-CoV-2 inferred from the ML viral phylogeny. Image Credit: https://www.biorxiv.org/content/10.1101/2020.12.14.422739v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Viral fitness
Viral fitness is defined differently at different scales. Within a host or a population, it determines the replication or growth rate and the transmission potential of the virus, respectively. Many factors shape viral fitness, both intrinsic and extrinsic, such as genetic and environmental, respectively. For instance, mutations can enhance viral fitness, as can extrinsic factors like the weather or host behavioral patterns.
Many mutations emerged in the first few months of the pandemic, including the D614G mutation in the receptor-binding domain (RBD) of the spike protein of SARS-CoV-2. This has rapidly become the dominant strain in every region where it was introduced. This has led many to attribute a fitness benefit to this mutation. Experimental studies also suggested better infectivity and increased replication rates with this mutation in vitro and in vivo.
However, scientists still disagree about the fitness benefit of the mutation at the population level. The effect of other mutations needs to be accounted for since they also change the baseline fitness of the virus. Also, an apparent fitness advantage may be nothing more than a benefit conferred by suitable weather and host behavior patterns that favor viral spread, independent of the effects of the mutation.
Study Details
The current study focused on viral phylogenetic tracing to estimate the effect of various factors on viral fitness. The researchers considered that a pathogen lineage with a higher fitness at the population level would have a higher frequency of transmission, and thus be encountered at a higher level over time. They will also develop more variants, show a higher rate of branching, and leave a trail of newer variants in population samples.
Taking this forward, they decided to use a multi-type birth-death (MTBD) analysis, in which viral fitness, in the form of the origin and cessation of different lineages, varies with the changing state or type of the lineage. Each such state may stand for the genotype or other viral feature that represents a distinctive trait.
Phylodynamic study
The study involved over 22,000 whole-genome SARS-CoV-2 sequences. For each variant with a frequency of over 0.5%, the researchers listed potential fitness predictors. Sixty-six of these had amino acid changes, and six had structural changes. Environmental predictors include the state/regional location of each sample. Each set of ancestral features provided a vector predicting viral fitness for that lineage.
To do this, they built a new method of birth-death modeling. Beginning from a reconstructed initial or ancestral state for every feature under consideration, they apply a fitness mapping function to convert this into the expected fitness. A predictive algorithm based on a combination of machine learning and likelihood-based statistical inferential methods is trained to learn this function from such a reconstructed phylogeny.
This approach helps avoid bias due to baseline differences. They were able to estimate the level of importance of different features contributing to the overall viral fitness. They call this decomposing or partitioning the difference in fitness between various viral lineages.
Transmission rates reflect local extrinsic factors
Even without using mutational data, the researchers found that implementing different transmission rates for different states and at different times brought the fit much closer to the modeled data.
The maximum likelihood estimate (MLE) of the base transmission rate was 0.16 overall. If 0.14 is taken as the rate of recovery or removal of cases, the basic reproduction number R0 becomes 1.15, vs the accepted estimate of 2-2.3. After accounting for the effects of space and time, the effective reproduction number Re becomes 1.05-5.84, with high variation between regions and time points.
Interestingly, the peak transmission rates were seen to be between February 1 and March 1, 2020, whereas the peak in reported cases falls much later. This difference, common to phylodynamic studies of this kind, has been attributed to large numbers of undetected cases and delays in case reporting. The estimated transmission was then found to fall and remain low until late summer, across the country.
Little impact of genetic factors
With this adjustment made for spatiotemporal differences in transmission rate, they estimated the effect of genetic variants on viral fitness.
They looked at sets of linked genetic variants, as is the case with the vast majority of cases. They found only one set had a harmful but weak effect on viral fitness. This may be because the study included only variants with a frequency above 0.5%, which would leave out most mutations with a strongly harmful effect.
Two independent variants had minor but significantly positive fitness effects, while two pairs of linked variants had significantly positive effects. Of interest, the set of D614G + nsp12 P323L variants had a very small beneficial effect on fitness.
How D614G became dominant
The study also suggests that the dominance of the D614G variant is simply due to the G614 strain being the first to be established in regions of the USA that were experiencing higher than average transmission rates – namely, the East Coast of the USA, especially New York and New Jersey. The enormous number of infections in the first wave of the pandemic more than accounts for the spike in the prevalence of the latter variant.
The baseline genetic fitness advantage was minor, about 4% on average. Both variants have a similar transmission rate. This type of comparison compensates for higher transmission or importation of the lineage into a region and allows the effects of any intrinsic fitness advantage to be seen.
Contribution of each factor
They then decomposed the total fitness variation between lineages into genetic, spatiotemporal, and random effects. Early on, fitness variation is almost completely due to the different transmission rates in different regions. At this point, all circulating strains have the same genetic background. Currently, half the variation is because of random effects, and half is due to genetic-cum-spatiotemporal effects.
Over time, a very small variation in fitness occurs, about 25% being due to genetic factors. It rises again in late summer, but this cannot be due to the spike D614G mutation because the D614 strain was already out of the door by then. Instead, the difference in fitness is probably because of the cumulative effect of multiple low-frequency variants, each with a modest effect on fitness, circulating in late summer.
What are the implications?
Up to this point in time genetic variants have contributed little to overall fitness variation in the SARS-CoV-2 viral population.”
Extrinsic factors like social distancing are responsible for the differences in transmission locally, and probably between different regions as well. However, limited data precludes a definitive conclusion.
The researchers use the analogy of “gene surfing”, where a mutation rapidly increases in frequency across a wide range of locations through individuals traveling far and wide from a focal location, to explain how the D614G spike variant rose to dominance.
“The gene surfing analogy captures the idea of how even a neutral mutation can be propelled to high frequencies across a range of spatial locations as a result of rapid population expansion.”
Further research in the way viral lineages change in accordance with alterations in human behavior is essential to quantify transmission rates from phylogenies. If validated, this approach would be extremely useful for assessing the impact of public health interventions on viral spread
The investigators emphasize that the D614G variant may enhance replication within the host, but may not increase infectivity or transmission rates between hosts. The logarithm of the viral load correlates with transmissibility but reaches saturation point rapidly. Moreover, the already high replication and high-affinity binding of the earlier variant does not favor further selection for a replication advantage.
The researchers caution that more efficacious mutations may well arise in the future, and their phylodynamic model would help sort out the important ones in terms of their transmission potential. The ability of this model to assess the relative fitness contribution of different components of viral fitness could be very helpful in the future, such as the effect of new mutations vs that of non-pharmaceutical interventions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources