As the coronavirus disease 2019 (COVID 19) pandemic continues to exert its influence over the world, rapid testing methods for the detection of RNA are urgently required for disease surveillance.
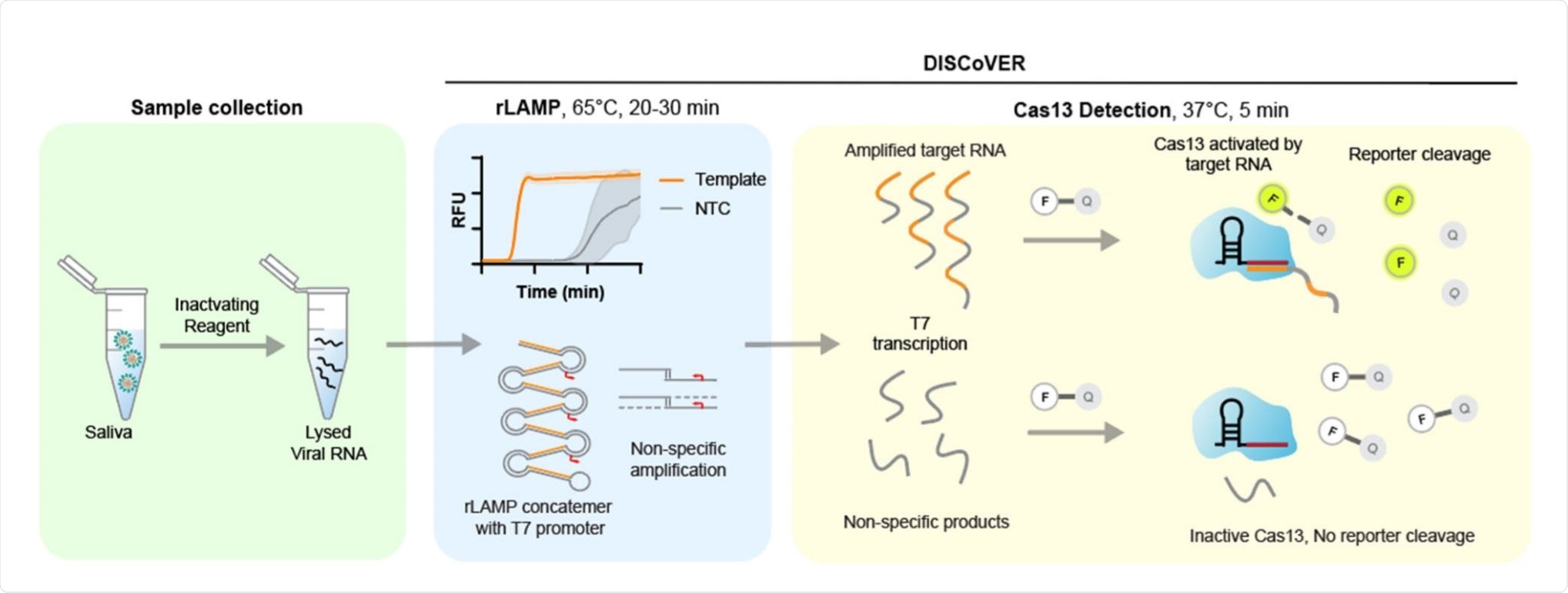
Schematic of DISCoVER sample to answer workflow. Image Credit:

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This test has high sensitivity and specificity and can be used with multiple CRISPR probes to detect a human gene or other pathogens along with the target virus. Called DISCoVER (DiagnosticS with Coronavirus Enzymatic Reporting), this is a SARS-CoV-2 test that does not require RNA extraction. In this, it is unlike other CRISPR-based tests like DETECTR and STOPCovid V2 that use LAMP and Cas12.
Secondly, DISCoVER avoids sampling of the upper airway, being based on saliva specimens. The use of saliva samples has been described, to avoid healthcare worker-patient contacts. These agree with nasopharyngeal swabs in 97% of cases when tested by reverse transcriptase-polymerase chain reaction (RT-qPCR).
Challenges in standard qPCR
Following infection with the severe acute respiratory syndrome coronavirus-1 (SARS-CoV-2), there is an asymptomatic phase where the virus replicates within the infected host cells and spreads to other parts of the lungs and the ciliated cells of the nasal airway. In the next phase, viral production becomes exponential and the individual becomes infectious, though often asymptomatic or presymptomatic.
Once the symptomatic phase sets in, the viral load in the upper airway is usually falling from its peak, and the rates of nucleic acid positivity are correspondingly lower. Since this is the phase when maximum testing rates are observed, many tests are likely to return false-positive results.
Moreover, standard tests such as quantitative PCR (qPCR) require sophisticated facilities, trained personnel, and take time. The long time required for a test result to be returned makes these tests unsuitable for POC diagnostics.
Need for rapid POC tests
The most efficient way to break the chain of transmission is by regular testing using rapid point-of-care (POC) testing at short intervals. Again, rapid testing could be used to identify positives at the community level, which are confirmed by more specific tests following referral.
Thirdly, having more than one or a few methods of testing could prevent bottlenecks in the diagnostic supply network by providing alternative sampling and testing kits.
Earlier researchers have developed assays that use qPCR without previous RNA extraction, with heat lysis of the sample. Chaotropic agents, RNAase inhibitors, and reducing agents have also been used.
Test Principles
CRISPR-based detection and isothermal amplification are methods that can be combined with direct lysis and saliva sampling for POC diagnostic testing. CRISPR-mediated detection of viral RNA depends on the use of guide RNA to activate Cas13 or Cas12, both of which are nucleases. These activated enzymes elicit nonspecific nucleases against single-stranded (ss) RNA or DNA, thus cleaving a bound reporter molecule and enabling its release. The detection of this reporter then provides the test readout.
The advantage of CRISPR-based testing is its high specificity. However, the use of Cas13 nucleases alone can delay attomolar sensitivity by up to two hours. To overcome this, they used loop-mediated isothermal amplification (LAMP), which is both rapid and highly sensitive, taking less than 20 minutes for detection at attomolar sensitivity.
With LAMP, the first viral RNA is reverse-transcribed to DNA using a combination of reverse transcriptase, a strand displacing DNA polymerase, and three primer pairs. The DNA is then subjected to LAMP.
In the current study, RNA polymerase promoters were integrated into the LAMP primers, so that the amplified DNA could be transcribed to single-stranded RNA, which is recognized by the Cas13 probe. This results in LAMP amplification to RNA or rLAMP. Since each LAMP product is transcribed by the RNA polymerase as soon as formed, the Cas13 signal peaks in <five minutes, respectively. Its limit of detection (LOD) is attomolar.
In the current experiment, they explored nine sets of LAMP primers that were directed at different regions across the genome of SARS-CoV-2 and found that all produced positive signals when they targeted genomic RNA at 100 copies/µL. Fluorescence peaked within 20 minutes, and for three of the sets, within 15 minutes.
This technique, however, requires correction for non-specific amplification, possibly the result of primer dimerization. This correction was provided by using a specific Cas13 probe to specifically recognize amplified nucleic acid, thus coupling sensitivity with specificity.
The researchers chose the N set 1 of primers, which recognizes the SARS-CoV-2 nucleocapsid (N) gene, both because it has a low time to the threshold and because its amplicon is large enough to fit in the Cas13 guide RNA. The N set 1 primer set has a LOD of 25 copies/µL.
What are the implications?
The current study thus describes an unusual combination of two amplification mechanisms, boosting the sensitivity of the test, with a Cas13 probe for specificity. LAMP amplification takes 20-30 minutes.
The use of common, inexpensive, and easily available reagents for direct lysis at room temperature simplifies the procedure. The researchers point out that the ease and comfort of saliva collection for COVID-19 testing will probably lead to higher testing rates and more frequent testing, including asymptomatic testing. This is because saliva samples have comparable viral titers vs upper airway swabs. However, the latter can also be used with the DISCoVER workflow.
The study validates the detection of SARS-CoV-2 on a saliva-based sample matrix, using a live virus, in parallel with a human gene as a control. Moreover, the ability to use different nucleic acids enables its adaptation for the detection of a variety of pathogens such as influenza A and B.
The use of saliva, RNA extraction-free detection, and the potential for multiplex target detection makes it an excellent candidate for development into a POC diagnostic test for COVID-19.
Further integration with a microfluidic platform and detection device will facilitate frequent testing for schools and workplaces as part of a robust infrastructure for pandemic surveillance.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Agrawal, S. et al. (2020). Rapid detection of SARS-CoV-2 with Cas13. medRxiv preprint. doi: https://doi.org/10.1101/2020.12.14.20247874. https://www.medrxiv.org/content/10.1101/2020.12.14.20247874v1
- Peer reviewed and published scientific report.
Chandrasekaran, Sita S., Shreeya Agrawal, Alison Fanton, Aditya R. Jangid, Bérénice Charrez, Arturo M. Escajeda, Sungmin Son, et al. 2022. “Rapid Detection of SARS-CoV-2 RNA in Saliva via Cas13.” Nature Biomedical Engineering, August, 1–13. https://doi.org/10.1038/s41551-022-00917-y. https://www.nature.com/articles/s41551-022-00917-y.