The onset of the current coronavirus disease 2019 (COVID-19) pandemic revealed the seventh known pathogenic human coronavirus, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).
Others include the endemic seasonal human coronaviruses, HCoVs, comprising two alphacoronaviruses, 229E and NL63, and two beta coronaviruses, OC43 and HKU1 (all of which cause common colds), the Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV), and the Severe Acute Respiratory Syndrome coronavirus (SARS-CoV).
The ongoing pandemic has been by far the most pathogenic and widespread of these viruses, with over 65 million documented infections and over 1.6 million deaths attributed to it, so far.
With no effective antiviral having been developed so far, physicians have turned to the time-tested approach of convalescent plasma (CP). Also, many monoclonal antibodies have been developed that target specific epitopes on the SARS-CoV-2 virus, to neutralize it. Early though it is, CP has received Emergency Use Authorization, and its benefit is supported by some preliminary data, especially when it contains high neutralizing titers.
The antibody response to the virus is polyclonal, with different patients showing varying titers of antibody, directed at different antigens, with varying kinetics of induction, the isotype usage, and the actual protection afforded by the antibodies.
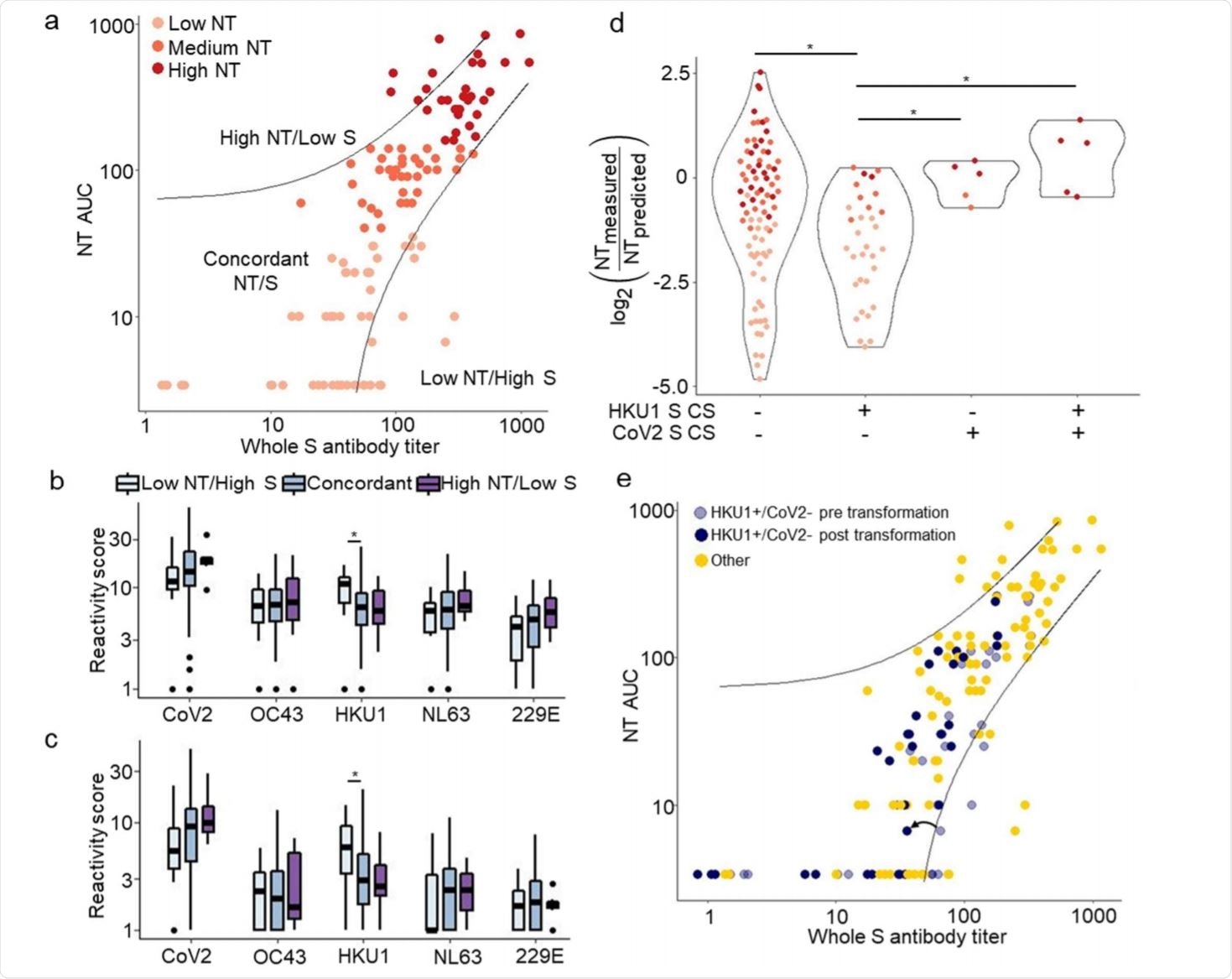
VirScan identifies features associated with discordance between whole spike titer and NT AUC. Image Credit: https://www.medrxiv.org/content/10.1101/2020.12.16.20248294v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Original antigenic sin
Pre-existing antibodies directed against the SARS-CoV-2 spike protein are present in a small percentage of the population. This may be due to the highly conserved nature of the sequences encoding the viral S2 subunit, which is essential for virus entry into the host cells.
Due to the shared nature of such sequences, the titers of such pre-existing antibodies may rise in response to SARS-CoV-2 infection, without a benefit to the patient in terms of neutralizing capacity or protection against the virus. This is called ‘original antigenic sin,” where the immune response against a pathogen or antigen primarily consists of antibodies or T cells formed during an earlier exposure to a related pathogen or antigen.
Study aim and details
The current study aimed to explore the hypothesis that this variation is determined by the pre-existing antibodies elicited by endemic HCoVs. The researchers made use of systems serologic testing and highly multiplexed epitope analysis, to determine the functional characteristics of CP, as well as the exact epitopes bound by various CoV antibodies. Their research was published in the preprint server medRxiv*.
They studied the antibodies in CP donated by 128 donors, at intervals of 13-67 days following confirmation of COVID-19 by reverse-transcriptase polymerase chain reaction (RT-PCR). The CP was donated for use in a clinical trial, and hence the titer of neutralizing antibodies (neutralizing titer, NT) was measured for each sample.
The researchers categorized the CP into high, medium, and low NT samples, where the area under the curve for NT was 160 or higher, 40-159, and less than 40, respectively. They also measured other functional characteristics, namely, the antibody-dependent cellular phagocytosis (ADCP), antibody-dependent cellular cytotoxicity (ADCC), and antibody-dependent complement deposition (ADCD) for each sample.
VirScan library
They used a large library of over 3.400 peptide epitopes, each 56 residues long, from an array of 230 COVID-19 samples and 190 samples pre-dating the COVID-19 onset. This was prepared using a multiplexed antibody profiling system, VirScan, which analyzed very large numbers of epitopes simultaneously. The library covers all seven human pathogenic coronaviruses, as well as three related bat coronaviruses.
Using the VirScan library, the researchers were able to identify the epitopes in each sample, while also doing a comparison on 87 samples from before the pandemic. Any region that showed 20% binding by an antibody was considered to be reactive. This turned up 27 regions of dominant reactivity, mostly corresponding to already known regions.
The NT AUC was not related to the extent of reactivity of these regions to specific peptides, except for one region overlapping with the spike receptor-binding domain (RBD) of the SARS-CoV-2. This region was more reactive in the high NT group.
This indicates that with individual peptides, the number of reactive epitopes is more important than the magnitude of binding by individual peptides in distinguishing low NT from high NT CP. With high NT samples, in 50% to 100% of cases, antibodies target three regions of the Spike (S) protein, one region of the membrane (M) protein, and four of the nucleocapsid (N) protein.
With medium and low NT CP, a lower frequency of recognition was observed. However, the researchers observed that low NT plasma was mostly reactive to S1/S2 cleavage site (CS) peptides from the seasonal beta coronaviruses.
High reactivity associated with high NT
High NT CP was associated with the highest reactivity for SARS-CoV-2 protein, but also higher total NL63 and OC43 reactivity, which indicates cross-reactivity. The greater the number of SARS-CoV-2 and NL63 dominant region epitopes that showed reactivity with the CP, the higher was the NT AUC.
Fusion peptides are highly targeted epitopes
Many of the dominant HCoV regions were found to show increased reactivity correlated with a rising NT AUC, such as the spike fusion peptide that is essential for cell entry. In public databases of SARS-CoV-2 proteins, this region is among the most frequently targeted epitopes, with high sequence sharing among the HCoVs. As a result, these epitopes induce a rise in cross-reactive HCoV/SARS-CoV-2 antibodies following infection with the latter.
Overall, the fusion peptides of the S2 subunit of the HCoVs and SARS-CoV-2 were the most reactive among all the NT groups for each coronavirus. Again, plasma that reacted with the SARS-CoV-2 fusion peptide was most likely to react with all other CoV fusion peptides. Even in pre-pandemic plasma samples, there was an 8% cross-reactivity with fusion peptide and HR2 peptide from SARS-CoV-2.
Peptides that reacted with the NT AUC also correlated with other functional characteristics of CP, as would be expected from their independent correlation.
Identification of high NT plasma
CP with increased potency is characterized by antibodies generated by multiple B cells, in response to multiple epitopes of SARS-CoV-2 and NL63, and by cross-reactivity to the fusion peptide of the former. When only HKU1 cleavage sites (CS) peptides were recognized, but not the CS peptides of SARS-CoV-2, neutralization activity was much lower compared to high NT CP. On the other hand, antibodies reactive for SARS-CoV-2 RBD rather than for HKU1 RBD correlates with high NT plasma.
Thus, the highly polyclonal nature of SARS-CoV-2 antibody responses defines a highly neutralizing response.
Individual reactivity predicts high NT
The peptide-antibody binding that was most likely to be associated with ADCC was from a part of the M region that activates T cell binding. Other functionalities of CP, such as ADCC, ADCP, and ADCD, are correlated with each other. The individual reactivity of each peptide may correlate with highly functional CP in one of two ways.
Firstly, the more severe the disease, the higher the NT, which may reflect in an individual increase in reactivity. Conversely, the greater the reactivity, the more effective the antibody response may be, and thus, the less severe the disease.
What are the implications?
The researchers suggest that much of the reactivity to HCoV peptides that are present in CP is due to the boosting of pre-existing HCoV antibodies by the SARS-CoV-2 infection – also called an anamnestic response – or due to actual cross-reactivity. As such, these HCoV antibodies could be useful as biomarkers, helping to identify plasma potency.
Moreover, their correlation with higher NT could indicate their protective role. The researchers observed that neutralization activity is most closely linked to the antibodies targeting the RBD, fusion peptide, and the CS peptide. The latter is likely to be important in neutralization, as therapeutic antibodies have triggered escape mutations in this region. Further study may reveal the interactions between anti-CS antibodies and the severity of COVID-19.
However, their analysis also showed that a stronger immune response to NL63 is linked to a stronger neutralizing response, as supported by earlier observations of milder disease with neutralizing antibodies to NL63. This could be because both this virus and SARS-CoV-2 use the ACE2 receptor.
The possibly protective nature of cross-reactive anti-HCoV/SARS-CoV-2 fusion peptides may support the utility of plasma stored in blood banks, containing anti-fusion peptide antibodies, in future outbreaks caused by coronaviruses.
As CP enters wider use, the reactivity of plasma to HKU1 CS is a potential marker that may help to identify plasma with high NT.
An understanding of the fine specificities of anti-coronavirus antibody repertoires may be applied to therapeutic plasma prioritization.”
The method described here can distinguish preferential epitope-recognizing antibodies from non-specific cross-reactivity, could help to differentiate original antigenic sin vs heterologous protective antibodies for other viruses as well, and to distinguish antibody responses to closely related viruses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources