The resurgence of the current coronavirus disease 2019 (COVID-19) pandemic has reinforced the need for rapid and reliable diagnosis of infection with its causative agent, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).
Proteinase K improves the performance of the heat inactivation method in RT-qPCR determinations of SARS-CoV-2. Image Credit: https://www.medrxiv.org/content/10.1101/2020.12.16.20248350v1.full.pdf

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The need to speed up RT-qPCR
While RT-qPCR is the gold standard for diagnosis of SARS-CoV-2 infection, the protocol followed at present requires RNA extraction, to produce the purified RNA substrate for the polymerase enzyme. Not only does this step add time, if performed manually, but it requires expensive reagents which can readily run out of stock under conditions of high demand, as at present. The alternative is the use of robots, which adds to the expense. The extraction of RNA from the virus also requires biosafety level 2 (BSL2) facilities.
All these obstacles create unwanted bottleneck in testing procedures. The author's comment, “In Argentina, a trained lab technician processes approximately 24 samples every 1.5 hours.” This is grossly inadequate to meet current requirements.
Alternative methods
Many researchers are working on methods that avoid this step. Several papers are describing the use of thermal inactivation with high-sensitivity detection of the short nucleocapsid regions of the viral RNA that survive the heating step intact. The various media that can be used with this technology are also described in earlier studies.
Another approach is the use of proteinase K (PK). This is a protease enzyme that breaks down RNases, thus preventing the degradation of RNA. The use of PK may thus help preserve the viral RNA intact during heat-inactivation protocols, but at the cost of some sensitivity.
Study details
The current study, which appeared as a preprint on the medRxiv server, describes a protocol that combines both PK and thermal inactivation, successfully determining the presence of SARS-CoV-2 RNA by RT-qPCR.
The test was conducted on nasopharyngeal swabs obtained during the screening of non-hospitalized symptomatic cases and their close contacts. The swabs were stored for up to 24 hours in saline and added to tubes pretreated with PK. This step minimizes the handling of potential virus-containing samples since these tubes are next handled only after inactivation.
Moreover, this step can be carried out outside the virus-handling area, allowing the workflow to be expanded significantly.
The researchers then heated the samples before proceeding to RT-qPCR for the detection of multiple viral genes, namely, N1 and, N2; N and ORF1; and N, E and RdRp, with human RP gene controls, in different assays. They also carried out infectivity assays using the same samples, to ensure inactivation.
The viral N, E, ORF1a, and ORF1b genes were amplified using loop-mediated isothermal amplification (LAMP). Finally, they calculated the efficiencies of the various PCR tests and analyzed the amplification curves.
Advantages of PK addition
The virus was found to be inactivated, as shown by the infectivity assays which showed no cytopathic effect or viral RNA detection by subsequent RT-qPCR of the supernatant.
They found that PK preincubation leads to a better PCR test performance following heat inactivation. The saline solution used in this case produced optimal results, unlike earlier reports. It is noteworthy that the CDC also recommends this transport medium.
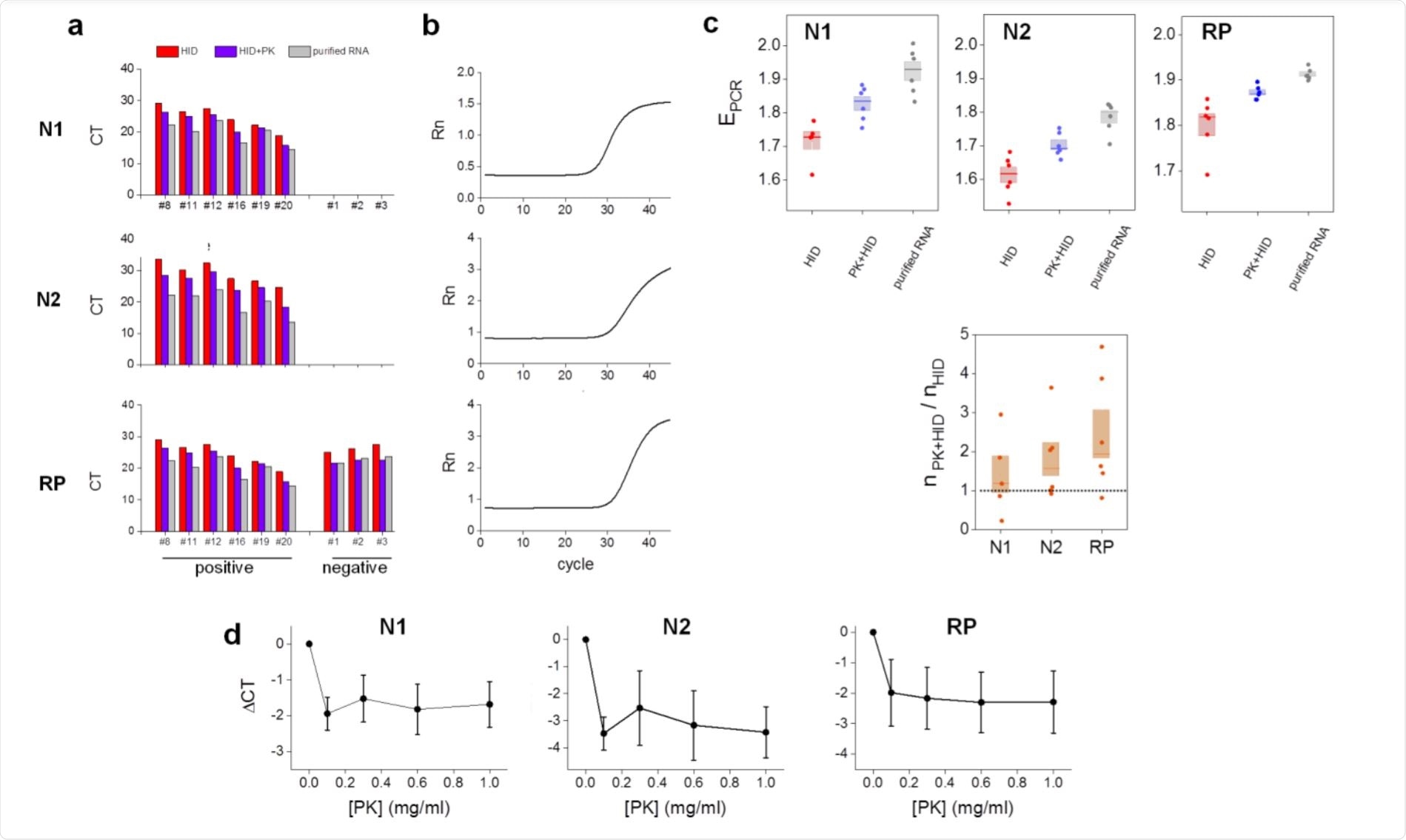
All samples were correctly identified as positive or negative after heat-inactivation or PK-heat inactivation protocols. The latter reduced the number of cycles required to detect the RNA relative to the former, as shown by lower cycle threshold values, which were more comparable to those determined using extracted RNA.
In other words, the efficiency was maximum for RNA extraction followed by RT-qPCR, then for PK-thermal inactivation, and lowest for heat-inactivation alone. Thus, the PK may reduce the concentration of unknown substances that interfere with the assay.
The N1 gene showed better efficiencies compared to N2, as reported by other researchers. The amplicon copy number with PK-heat inactivation samples compared to that with heat-inactivation alone was higher for N2 and RP, perhaps because PK prevents these regions also from being broken down by RNase.
The addition of PK increases the dilution, which could contribute to the overall efficiency of detection. At higher volumes, again, detection sensitivity rises in samples with low titers of the virus. However, it was independent of PK concentrations within the range of 0.1 to 1 mg/ml, allowing for still cheaper tests and also increasing the reliability of the test results.
High detection sensitivity
When detection of only the N1 and the human RP gene control was directly compared with purified RNA samples, the PK-heat inactivated extraction-free samples showed 100% sensitivity and specificity, with very similar Ct values, showing that the new method accurately reflects the presence of the virus.
The researchers also investigated the use of this PK-heat inactivation method with multiplexed kits that detect other genes in multiple reactions. For instance, DisCoVery (targeting N and ORF1ab genes) and Genefinder (targeting N, E, and RdRp genes) were used here, and are commonly used in clinical situations.
The findings show that there is no variation in the detection of these samples prepared by either method, even with Ct values over 35. Despite the weaker detection of E and RdRp with the Genefinder compared to N, the test was still very sensitive for viral RNA detection.
They found similar amplification efficiencies for N, ORF1ab, and RP genes by both methods. These samples can be stored at 4°C following PK-heat inactivation for up to 20 hours, or freeze-thawed, without any change in Ct values. Validation assays showed an accuracy of 99%, with sensitivity and specificity both being 99%.
What are the implications?
The use of PK followed by heat inactivation is a reliable protocol for RT-qPCR detection of SARS-CoV-2 without RNA extraction. This method can also be combined with LAMP kits, which are in widespread use due to their convenience and cheapness.
The simplicity and relatively short period required for the protocol and the full-availability and low price of the required reagents place PK+HID as a cheap, simple, and fast alternative to traditional RNA extraction protocols.”
If validated, this could provide an alternative testing pathway that is flexible for multiplex testing, inexpensive, and accurate. This could result in a marked improvement in testing capacity in low-resource settings.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources