Researchers in the UK have conducted a comprehensive epidemiologic analysis of the new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant in an effort to learn more about the expanded range and frequency of coronavirus disease 2019 (COVID-19) cases in England.
The SARS-CoV-2 lineage B.1.1.7, which was first detected in the UK in November 2020, has been designated Variant of Concern 202012/01 (VOC) by Public Health England.
The team’s analysis of VOC whole genomes identified an increased frequency in the variant that is consistent with a transmission advantage over other SARS-CoV-2 variants in the UK.
The estimated difference in reproduction number (number of new infections arising from a single case of infection) between the VOC and non-VOC lineages ranged between 0.4 and 0.7.
The team – from Imperial College London, the University of Edinburgh, Public Health England, the Wellcome Sanger Institute, and the University of Birmingham – warns that this substantial transmission advantage will pose major challenges for the ongoing control of COVID-19 in the UK and elsewhere over the coming months.
Importantly, the researchers found evidence that while non-pharmaceutical interventions (NPIs) were sufficient to control non-VOC lineages during the November 2020 lockdown in England, they were not sufficient to control the VOC.
Neil Ferguson and colleagues warn that social distancing measures will need to be more stringent and that policy decisions will need to be informed by urgent epidemiological investigations into the new variant.
A pre-print version of the paper is available on the medRxiv* server while the article undergoes peer review.
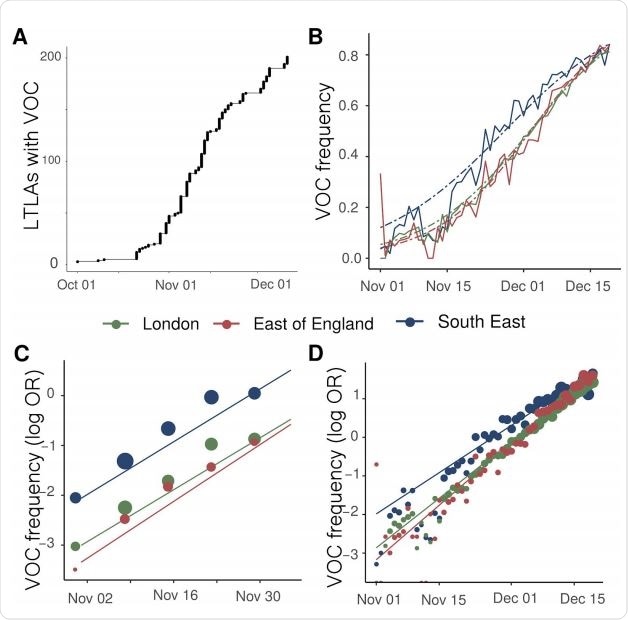
Expansion and growth of the VOC 202012/01 lineage. A) The number of UK LTLAs reporting at least one sampled VOC genome. B) Empirical (solid) and estimated (dash) frequency of TPR-adjusted SGTF in three regions of England. C) Empirical (points) and estimated (line) frequency (log odds) of VOC inferred from genomic data by epidemiological week. D) Empirical (points) and estimated (line) frequency (log odds) of SGTF based on the same data as B.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The VOC possesses advantageous mutations
As of 20th December 2020, the regions in England with the highest number of confirmed cases of the new variant were London, the South East, and the East of England.
The variant possesses a large number of non-synonymous substitutions that are of immunologic significance because they increase host cell binding and infectivity.
One deletion mutation that occurs at positions 69 and 70 (Δ69-70) on the viral spike protein (which binds host cells) has been associated with the failure of diagnostic PCR tests that probe this protein.
“The absence of detection of the S gene target in an otherwise positive PCR test increasingly appears to be a highly specific marker for the B.1.1.7 lineage,” writes Ferguson and colleagues. “Surveillance data from national community testing (‘Pillar 2’) showed a rapid increase in S-gene target failures (SGTF) in PCR testing for SARS-CoV-2 in November and December 2020.”
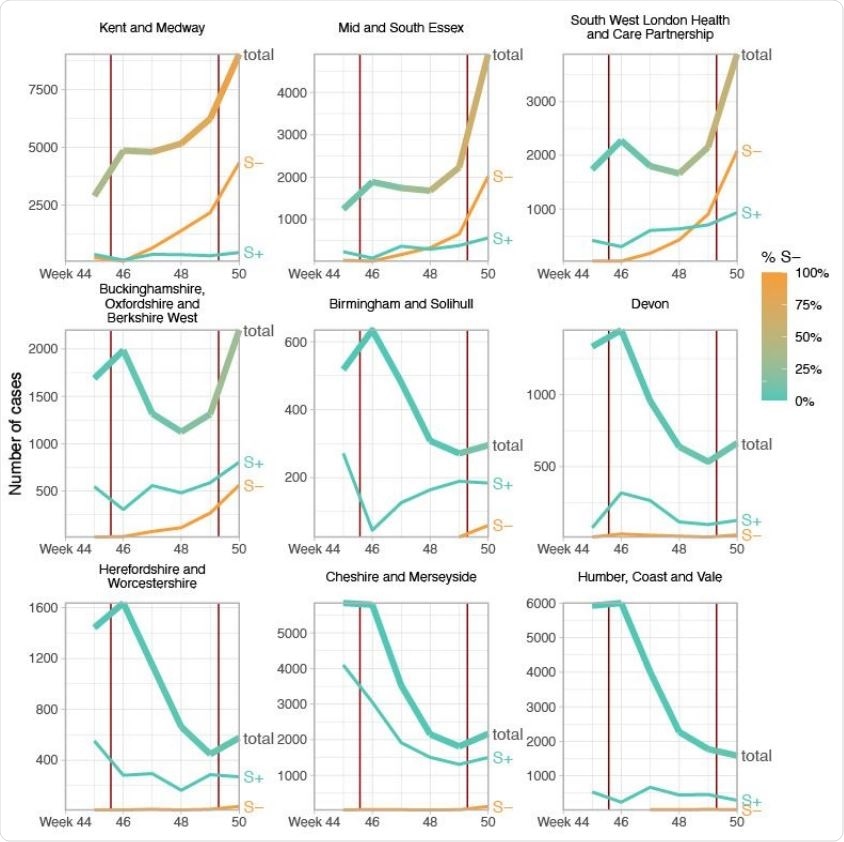
Case trends in a subset of NHS STP areas. Total cases reported are shown as a thick line. A subset of these - those tested in the 3 largest “Lighthouse” laboratories - were tested for SGTF. The total cases line is coloured according to percentage S- among those tested. Counts of S+ and S- reported via the PHE SGSS system are shown by the thin lines. The dates of the second lockdown are indicated by the vertical red lines. Nine representative NHS STP areas from all regions of England are ordered by decreasing percentage S- in the most recent week of data. Raw SGTF data are shown here (not adjusted for TPR), so S- cases in earlier weeks include other non-VOC lineages, especially outside the East and South East of England. Plots for all STP areas are shown in Figure S3.
What did the study involve?
The researchers used various statistical approaches to assess the association between the intensity of SARS-CoV-2 transmission and the frequency of the VOC, B.1.1.7, between November and December 2020 across different regions of the UK.
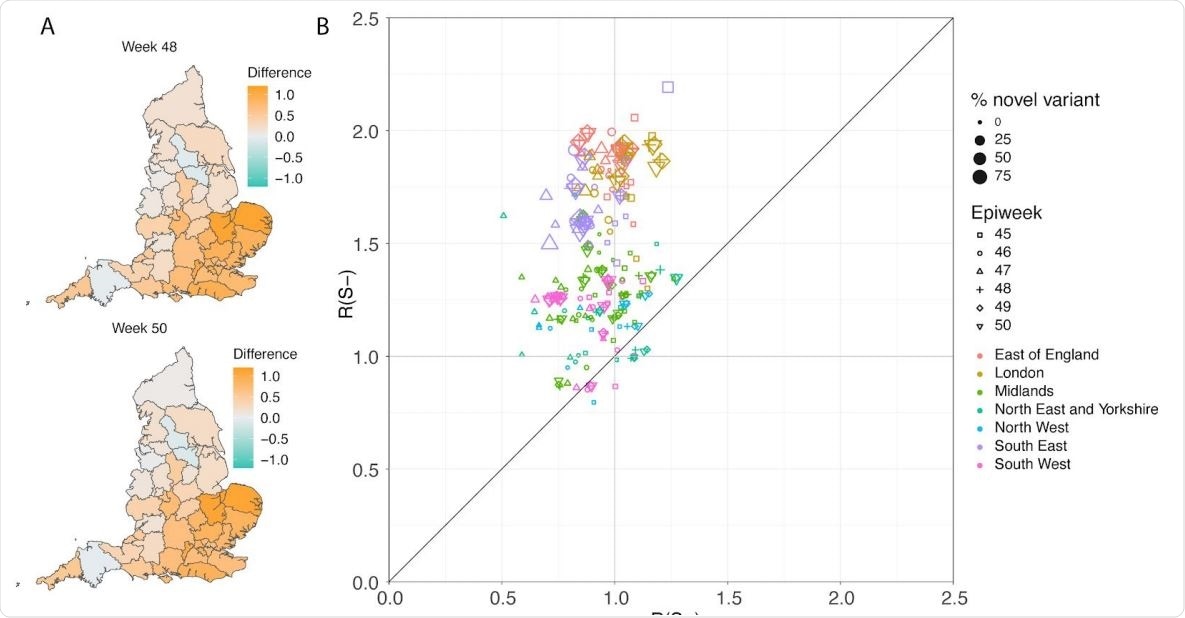
(A) Map of the difference in median Rt estimates for VOC and non-VOC variants for all STPs for weeks 48 and week 50. (B) Scatterplot of the reproduction numbers of VOC (S-) and non-VOC (S+) by STP and week. Point size indicates frequency of the VOC, while shape and colour signify week and NHS region, respectively.
The team’s analysis of whole-genome sequence data collected from community-based diagnostic testing showed that VOC sequences were widely distributed across 199 lower tier local authorities (LTLAs) in England. The sequences were most concentrated in the South East (n=875), London (n=636), and East of England (n=293).
“While rapid growth of the variant was first observed in the South East, similar growth patterns are observed later in London, East of England, and now more generally across England,” writes Ferguson and colleagues.
The researchers say the growth of the VOC lineage is consistent with it having a transmission advantage over other circulating SARS-CoV-2 lineages in England.
Next, the team investigated time trends in the proportion of PCR tests exhibiting SGTF across 275,000 test results.
The changes in VOC frequency inferred from the genetic data closely corresponded with the changes inferred by SGTFs in community-based diagnostic PCR testing.
A shift towards the under 20s
By examining growth trends in SGTF and non-SGTF case numbers at the local area level across England, the researchers showed that the VOC exhibits higher transmissibility compared with non-VOC lineages. The SGTF data also indicated a small but significant shift towards those aged under 20 years being more affected by the VOC than non-VOC variants, even after adjusting for variation by week and region.
Next, the team performed regression analyses to explore the effect of VOC frequency on the overall time-varying reproduction number. The analyses were conducted at two different spatial scales – LTLAs and NHS England Sustainability and Transformation Plan areas (a subdivision of NHS Regions).
Across almost every area analyzed, the researchers observed a very clear visual association between SGTF frequency and epidemic growth of the VOC.
What did the researchers conclude?
Ferguson and colleagues say that across all of the analyses conducted, the VOC was found to have a strong transmission advantage, with the estimated difference in reproduction numbers between the VOC and non-VOC lineages ranging from 0.4 to 0.7.
“The substantial transmission advantage we have estimated the VOC to have over prior viral lineages poses major challenges for ongoing control of COVID-19 in the UK and elsewhere in the coming months,” writes the team.
“Critically, we find evidence that non-pharmaceutical interventions (NPIs) were sufficient to control non-VOC lineages to reproduction numbers below 1 during the November 2020 lockdown in England, but that at the same time, the NPIs were insufficient to control the VOC.”
The researchers say social distancing measures will need to be stricter than they would otherwise have been.
“These policy questions will be informed by the ongoing urgent epidemiological investigation into this variant, most notably examining evidence for any changes in severity, but also giving more nuanced understanding into transmissibility changes,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ferguson N, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv, 2020. doi: https://doi.org/10.1101/2020.12.30.20249034, https://www.medrxiv.org/content/10.1101/2020.12.30.20249034v2
- Peer reviewed and published scientific report.
Volz, Erik, Swapnil Mishra, Meera Chand, Jeffrey C. Barrett, Robert Johnson, Lily Geidelberg, Wes R. Hinsley, et al. 2021. “Assessing Transmissibility of SARS-CoV-2 Lineage B.1.1.7 in England.” Nature 593 (7858): 266–69. https://doi.org/10.1038/s41586-021-03470-x. https://www.nature.com/articles/s41586-021-03470-x.