Since the onset of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, which is sweeping the whole world, scientists have been sequencing the RNA genome of the virus.
The virus’s genome has undergone numerous and frequent changes, which have led to first one, and then another viral strain becoming dominant in some countries and now the world over. A new study published on the preprint server bioRxiv* describes the results of genomic sequencing of the virus, which may help trace the routes of transmission of the virus from person to person, and from country to country.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Original sequencing results showed that the viral genome was very similar to two other SARS-like coronaviruses originally derived from bats, namely, RaTG13 and RmYN02, both from Yunnan province of China. Tracing SARS-CoV-2’s descent confirmed its close identity with these viruses and also showed that it to be different from the earlier coronaviruses, the severe acute respiratory syndrome coronavirus (SARS-CoV), and the Middle East respiratory syndrome coronavirus (MERS-CoV). Some scientists think that the current virus might be descended from one of the bat variants through an intermediate host.
The current study focused on over 4,000 full-length sequences of the viral genome retrieved from the Global Initiative for Sharing All Influenza Data (GISAID) EpiFlu database, for the most part. However, 11 came from a Chinese database. The sequences were uploaded over 14 weeks from the start of the Wuhan outbreak. The researchers looked at the mutations to characterize the genotypes.
They also analyzed another ~2,61,000 genomes collected globally over the 12 months since the pandemic began. This comprises all the genomes in the database.
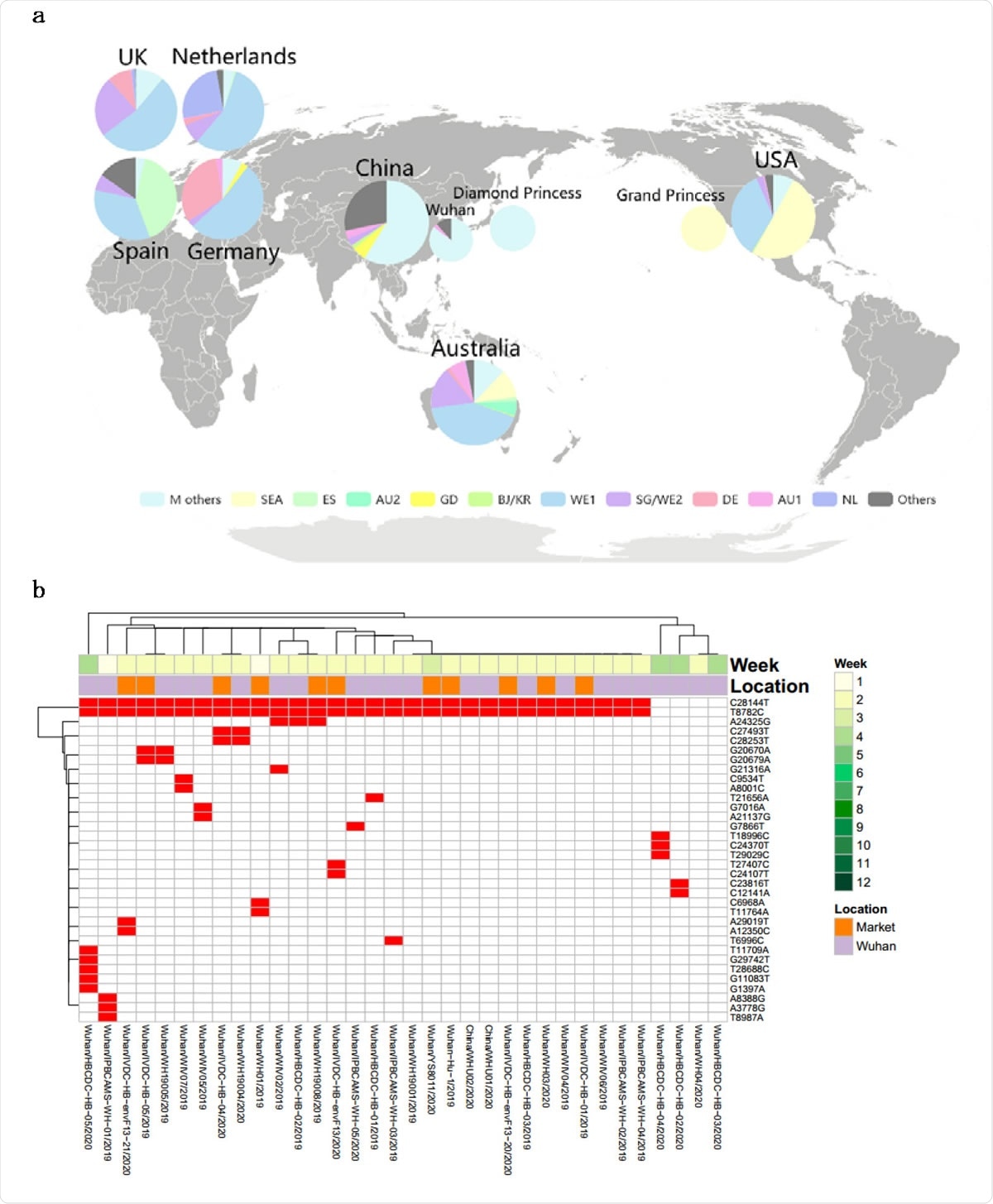
Major genotypes identified in the study. (a) Unsupervised mutaion clustering of all samples. Mutations concurently called from at least 5 samples were included. 11 distinctive major mutation profiles were identified based on clustering tree branches and were named mainly based on the geographic locations where a certain genotype was initially or mainly reported from. The two-letter ISO country codes were used to indicate the countries associated with the mutation profiles (as shown at lower color bar). The upper color bar demonstrates genotypic homogeneity within each clustering tree branch. (b) Pedigree chart of major genotypes. In combination of mutation clustering and available epidemiologic information, 11 distinctive main genotypes were characterized and the pedigree chart demonstrated the relationship of each genotype. The genotypes from Diamond Princess and Grand Princess derived from M type and SEA type respectively, were indicated with dashed arrows.
Superspreaders introduced distinct genotypes
The researchers were able to identify distinct genotypes based on how commonplace certain mutations were. This helped trace superspreaders since they shaped the pandemic to a large extent. These individuals passed on specific genotypes with certain highly prevalent mutations. A single introduction of such genotypes led to an outbreak of infection, increasing evolution with spread.
Among these superspreader genomic sequences, the M type variant accounted for over 80% of the sequences in the study. From their estimates of the expected rate of substitutions in the genome, the researchers conclude that this can be called a true founder effect rather than being due to many identical superspreading genomes.
Six descendant genotypes
They found six descendant genotypes that were directly derived from the ancestral strain via characteristic mutations. The most prevalent genotype among these was the WE1 type, defined by four mutations. Three of the four defining mutations of the WE1 strain were found in three early samples collected in January 2020. Among WE1 genomes, 70% came from Western Europe (the UK, Iceland, Belgium, France, and the Netherlands, perhaps by traffic across the borders. It also made up ~35% of cases in the US.
The SEA type is the most common in the USA, however, but was isolated from three other countries, namely, Australia, Canada, and Iceland, indicating that cases from the USA had been imported there. This is also called the Washington State outbreak clade. The other four descendant genotypes were confined regionally.
Looking at the mutations of the infective strains in four areas, the researchers concluded that the M type spread from Wuhan to other regions in China before the Wuhan lockdown. Examining 34 sequences from early Wuhan cases showed two clusters, 30 belonging to the M type, but with extensive diversity. The remaining four formed another co-circulating cluster. Thus, at this early stage, there were 18 different genotypes among the 34 sequences.
In the USA, the prevalent strains belonged to the non-M types, probably from 12 cases imported from the Hubei province. These, in fact, were the earliest cases reported in the USA, with each showing a distinct genotype.
Half the US cases were SEA type, while ~35% were WE1. It indicates that the USA “endured the first wave of case importation from China and the second wave from Europe, which is consistent with the recent COVID-19 study of Washington State.” Among the 32 patients on the two cruise ships, the Grand Princess and the Diamond Princess, there were 25 different genotypes. This indicates that the virus mutates rapidly and extensively during person-to-person transmission.
Strain of origin algorithm more accurate
The researchers developed a Strain of Origin (SOO) algorithm to match each genotype to its genome by mutational profile. When compared with mutation clustering, this approach showed a 90% agreement. “SOO represents a more accurate approach to define genotypes as it only takes into consideration the specific mutations of the particular genotypes with little influence from the rest random mutations.”
Using the same approach, they found that three of the top four GISAID clades were descendants of WE1. They estimated that one of three nucleotides in the viral RNA had undergone mutation over the 12 months of the pandemic.
The story of the pandemic
They analyzed the top 100 mutations and generated a lineage-based pedigree chart. This story begins with a putative first case, supposed to be a patient with an ancestral SARS-CoV-2 genotype, and postulated to be present on November 17, 2019. This led to more infections. By January 1, 2020, the Huanan market was locked down, and 19 M type genome samples were documented.
However, the M type had already been incubating in the market for weeks, which accounts for the vast majority of genomes belonging to the M type at this time. With the expansion of the outbreak into Wuhan city at large, the city was locked down on January 23, 2020, with 80% of the viral genomes being of the M type. However, the Spring Festival had already prompted extensive travel to and from Wuhan, leading to the Chinese and then the global outbreak of COVID-19.
By April 7, 2020, more than 80% of cases worldwide were M type, but in September, 70% belonged to WE1, in three clades, namely, GR, G, and GH. The rise in M type continued, making up ~98% of cases by December 25, with almost 90% being caused by WE1 strains.
The importance of the study
The researchers conclude that beginning with a single superspreader incident, the M type exploded over the world, following a few initial weeks when it passed unrecognized and uncontrolled. The M type acquired two concurrent mutations first, with another four defining mutations that led to the emergence of WE1 strains, and finally, another three that led to the WE1.1 strain. The rate of viral evolution, at ~27 substitutions per year, is not unusual, but the mechanism is still unclear.
Of the two new mutant strains attracting much attention, namely, the D614G point mutation and the N501Y mutation in the receptor-binding domain, both in the spike protein, are thought to be highly transmissible compared to the ancestral strain. The former was first documented in Western Europe in February 2020 and now makes up ~90% of strains, while the latter was first found in New York City on April 21, 2020, and makes up only 0.02% of cases.
The researchers caution that this study does not allow neutral mutations to be distinguished from adaptive mutations. However, they say, the genotypes of this virus serve as unique identifiers, helping to trace the transmission pattern of the virus backwards and reveal its pattern of expansion forwards. They point out that their algorithm can help correlate the viral genome to the genotype, if known, very accurately, aligning with its mutation profile. New genotypes can also be incorporated into it as they arise to further improve its performance.
“This study not only provides an unprecedented window into the global transmission trajectory of SARS-CoV-2 in the early phase, but also reveals the subsequent expansion patterns of the pandemic.”
Large-scale genomic sequencing could therefore be very helpful in tracing such patterns in an outbreak caused by a new pathogen, helping to develop rapid countermeasures to contain it in the worst-hit areas.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources