Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that caused the coronavirus disease 2019 (COVID-19) pandemic, is still spreading and mutating. Scientists are working on therapeutics to control the virus. An interesting new study published on the bioRxiv* preprint server describes a new platform that allows a high-throughput workflow to generate matched human lung buds in the tens of thousands. This allows researchers to study SARS-CoV-2 infection and therapeutics in lung tissue that mirrors the key aspects of human lung development.
Infection with SARS-CoV-2 may occur via the nose, or the airways of the lungs, and the alveoli. The virus seems to target the lung, in particular, with acute lung injury and pneumonia-associated complications are the most common complications of severe COVID-19. The virus attacks the ciliated cells and type II pneumocytes of the lungs via the angiotensin-converting enzyme 2 (ACE2) receptor as well as other enzymes that act as cofactors in viral entry.
However, the manner in which the virus damages lung tissue is still not clear. At present, most research on SARS-CoV-2 is carried out in human primary lung tissue. However, these in vitro systems are unpredictable and the cells vary widely, depending on the tissue donor’s genetic and phenotypic profile. These differences are important in their impact on viral replication and clinical impact.
One alternative to this is the use of differentiated human pluripotent stem cells (hPSCs) into lung airway and alveolar cells, while other researchers have developed lung cells from hPSCs from organoids to identify and test small molecules that inhibit SARS-CoV-2. However, these models do not show the tissue organization found in developing and adult lung tissue, resulting from embryonal signaling pathways that guide lung development. Moreover, lung cells take 1-3 months to differentiate from hPSCs.
Synthetic human lung buds
The current study aimed to produce a platform to generate synthetic embryonic lung-like tissues. These are formed from hPSCs, using their ability to self-organize when cultured in a specific geometry on micropattern chips. They used specific activators to generate thousands of lung progenitor cells, arranged in colonies, on a single micropattern chip.
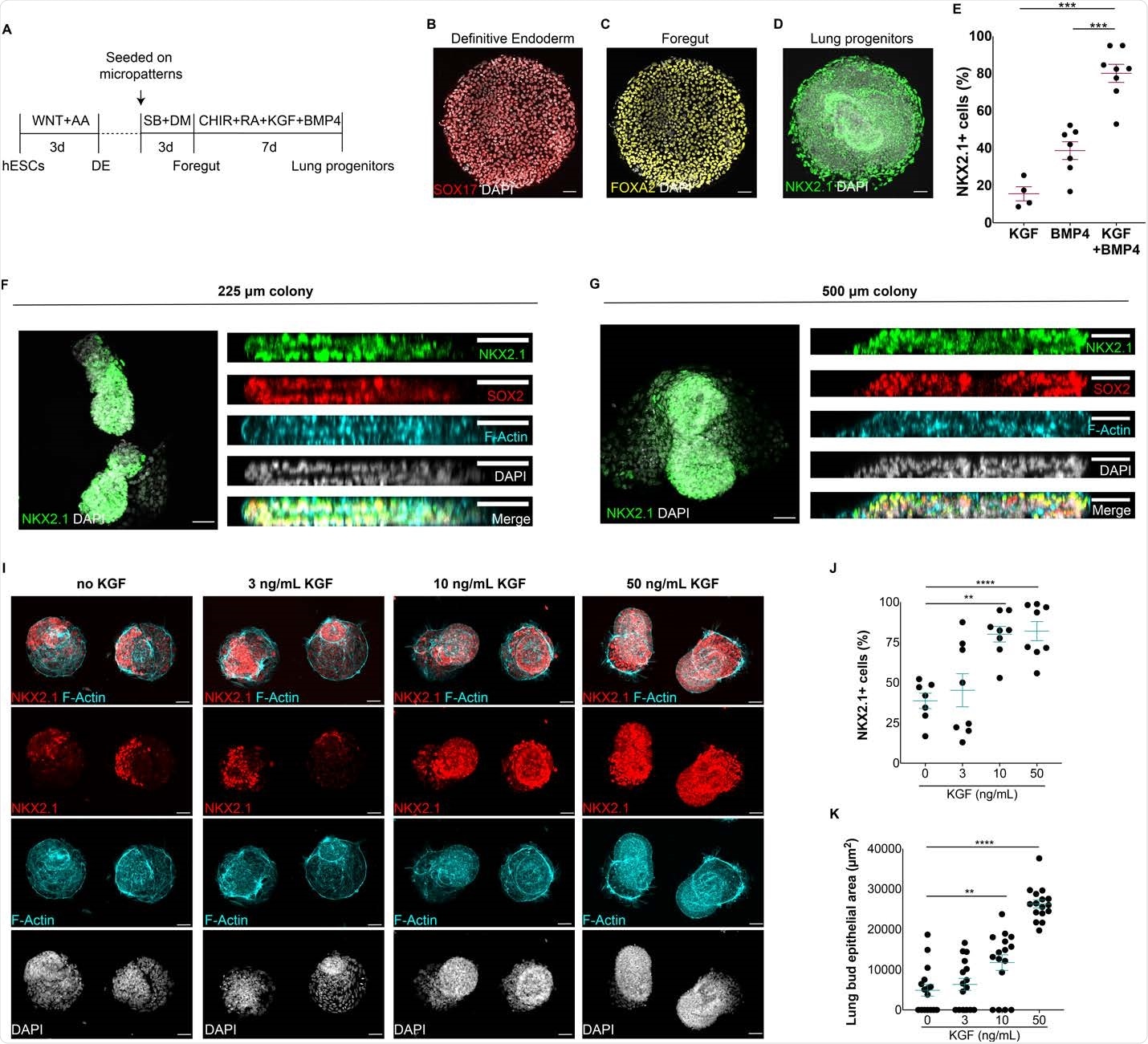
Generation of self-organized epithelial lung buds on confined geometries. A) Protocol for the generation of synthetic lung buds on confined geometries in micropatterns. B-D) Generation of SOX17+ endoderm cells (B), FOXA2+ anterior endoderm cells (C) and NKX2.1+ multipotent lung progenitors (D) at the end of definitive endoderm (DE), foregut and lung progenitor induction stages, respectively. E) Proportion of NKX2.1+ lung progenitors upon modulation with BMP4, KGF or BMP4/KGF for 7 days. F-G) Top and side views of 3D epithelial buds containing NKX2.1+ multipotent lung progenitors in 225 and 500 μm colonies. I) Micropattern colonies containing NKX2.1+ epithelial buds at increasing doses of KGF. J) Proportion of NKX2.1+ progenitor cells at increasing doses of KGF. K) Quantification of lung bud epithelial area in micropattern colonies at varying doses of KGF. (**p<0.01,***p<.001, ****p<0.0001, Dunnett’s multiple comparison test; scale bar, 50 μm)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
They found that not only did these synthetic human lung buds show early differentiation parameters, but they organized themselves into human lung buds. They were also able to discover some crucial steps in early differentiation and uncover novel relationships among alveolar cells.
They found that the virus preferentially infects these lung buds, both ciliated cells, and type II alveolar cells. The latter are more susceptible because they have a higher level of ACE2 and TMPRSS2, as well as furin expression, all of which are essential for SARS-CoV-2 infection. They also found suggestions that dividing alveolar stem cells are also more vulnerable to the virus.
The researchers measured infection at 48 and 96 hours from the start and found that infected cells kept increasing over time in all types of cells, suggesting that viral spread is happening within the synthetic human lung buds. They also found apoptosis occurring in alveolar and airway cells. They were thus able to use a single micropattern chip growing hundreds of synthetic human lung buds to track susceptibility to infection, identify the cell types that are susceptible, evaluate viral transmission between cells, visualize the cytopathic effects in many hundreds of synthetic lung buds on and thus follow the whole viral lifecycle.
They then did a screening for therapeutics that had shown in vitro efficacy in binding assays and in cell lines. They tested neutralizing antibodies in human convalescent plasma, for their efficacy in preventing infection over a range of dilutions. They found that the results were comparable to those obtained using cell lines. Again, they discovered that the neutralizing antibodies could also inhibit the spread of the virus through the organoid even after infection has occurred. This supports the potential of these drugs as therapeutics, functioning as biologic inhibitors of SARS-CoV-2 infection and cell-to-cell transmission.
What are the implications?
The scientists not only demonstrated that lung stem cells can self-organize when cultured in confined geometries but that these organoids repeat the same pathway that fetal lung progenitors go through during lung development. This could be of great benefit in research on lung disease and regenerative medicine that involves lung tissue models by providing rapid and unlimited access to synthetic genetically matched human organoid lung tissue.
Variations in disease susceptibility between different cells, and differences in the viral impact on different cell types, can also be studied quantitatively using this platform. In fact, this model allowed the scientists to identify some important types of cells that are otherwise difficult to study. It could also lead to a better understanding of why SARS-CoV-2 is more pathogenic than other coronaviruses.
Moreover, these findings show new types of cells and pathways of cell development in fetal life, which could help understand how the human lung differentiates. This could shed light on the origins of lung cancer and other diseases as well.
“We expect our synthetic human lung model to be transformative for the elucidation of the molecular basis of respiratory infections and lung diseases for which there are currently no treatments. This study also highlights the potential of using synthetic lungs to identify novel therapeutics that block infection by SARS-CoV-2, endemic coronaviruses as well as other respiratory viruses.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources