As the coronavirus disease 2019 (COVID-19) pandemic continues to progress, scientists have been working hard to find new ways to counteract the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) threat.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Human naïve antibodies contain anti-SARS-CoV-2 RBD antibodies
The virus elicits neutralizing antibodies that prevent viral entry into the host cells. Most of these antibodies are directed against the viral spike glycoprotein, specifically its receptor-binding domain (RBD), a small immunodominant region. Antibodies to the RBD are of several types, each type targeting a different epitope, though there is a degree of overlap. This indicates that neutralization can occur via multiple mechanisms occurring via inhibition of viral entry at more than one site on the RBD.
The majority of RBD antibodies described so far are surprisingly similar to their germline precursors, a state called low somatic hypermutation. This means that not only is it easy to produce anti-SARS-CoV-2 RBD antibodies, it makes it convenient for researchers to use naïve antibody libraries to pick out novel therapeutic antibodies and to compare antibody clones selected by their ability to bind the viral antigens in vitro with those that arise during natural infection or vaccination.
The researchers used yeast display techniques to isolate three novel neutralizing antibodies from one such library. Of these antibodies, ABP18 blocks RBD-ACE2 binding, while two (NBP10 and NBP11) do not. Thus, the technique proved to be capable of differentiating antibodies that blocked ACE2 and those that did not.
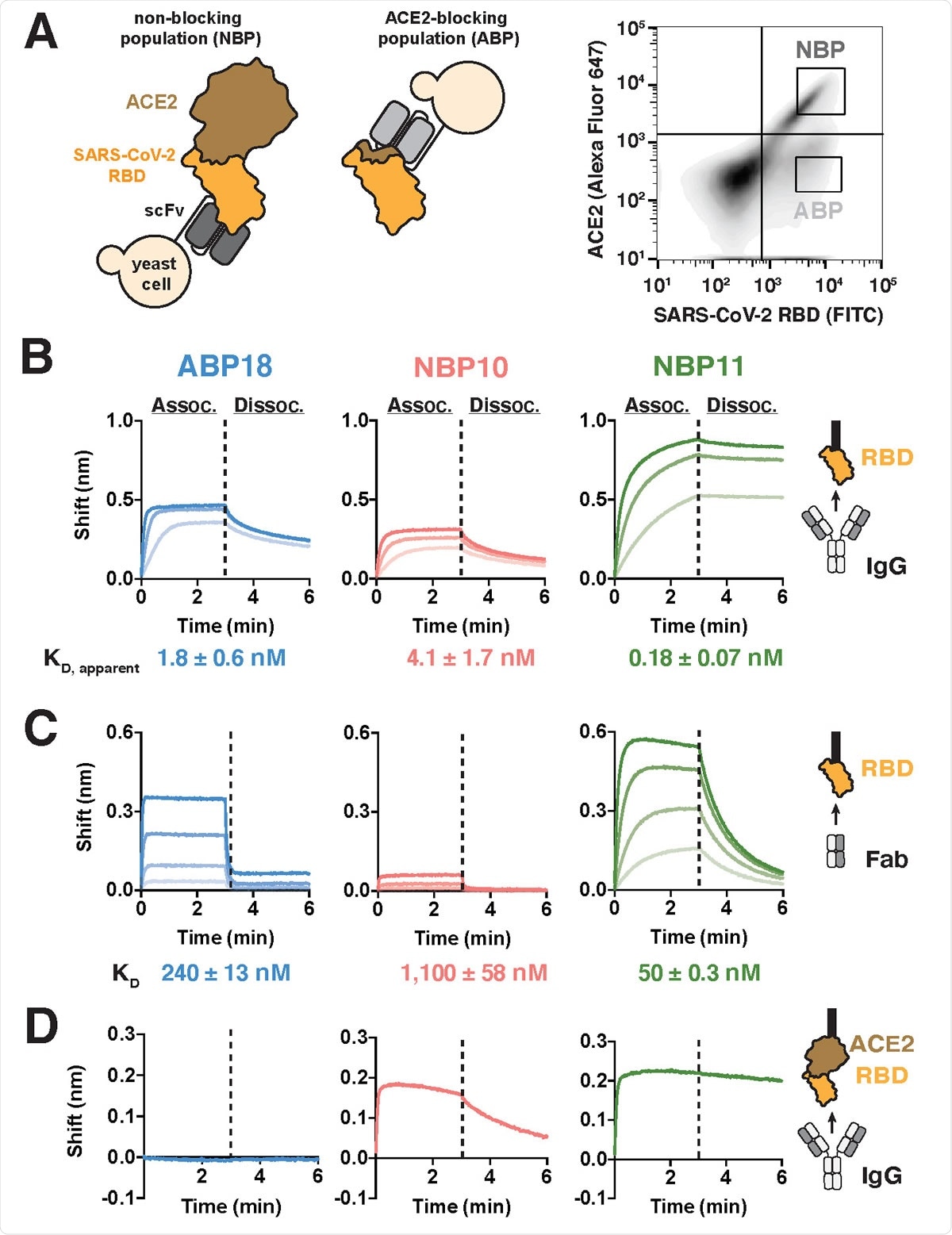
A human naïve antibody library yields antibodies targeting the SARS-CoV-2 RBD (A) Left, selection scheme to isolate an ACE2-blocking population (ABP) and a nonblocking population (NBP) of antibodies. Right, flow cytometry panel of yeast display library with gates used to isolate the ABP and NBP. (B-C) Binding to SARS-CoV-2 RBD-coated BLI biosensors show substantial differences between (B) IgG antibodies (100, 33.3, and 11.1 nM) and (C) Fab antibody fragments (ABP18/NBP11: 500, 167, 56, 18 nM; NBP10: 720, 240, 80, 27 nM). Higher antibody concentrations are indicated with thicker lines. (D) Binding of 100 nM IgG antibodies to BLI biosensors coated with ACE2:RBD complex shows that NBP10 and NBP11 IgG retain binding while ABP18 shows no binding, consistent with ACE2-blocking activity.
Anti-RBD antibodies neutralize spike-expressing pseudovirus
All three antibodies blocked the binding of a spike-bearing pseudovirus with a low half-maximal inhibitory concentration (IC50). The affinities of the non-RBD-binding antibodies were much more potent than their IC50 values, by 20-50 times, perhaps because they use a different mechanism of neutralization or because of different levels of accessibility of the epitopes concerned in the full-length spike protein.
All these antibodies have more potent neutralizing activity than their Fab binding affinity for the viral antigen. This suggests that the avidity of an antibody is important in viral neutralization.
Novel anti-RBD neutralizing antibodies engage distinct epitopes from natural antibodies
Using another technique called biolayer interferometry (BLI), it became apparent that the epitopes of all three antibodies are different from each other while overlapping with epitopes shared by natural antibodies. Each of the antibodies strongly competed with one or more natural antibodies, and each natural antibody competitively inhibited one or more other antibodies.
While NBP10 and NBP11 showed very similar binding competition, which suggests that their epitopes are similar and are encoded by a common heavy chain germline, IGHV5-51, they compete with one known patient-derived ACE2-inhibiting antibody COVA2-05 but not the others, CR3022 or EY6A.
CR3022 and EY6A two share many epitopes. Yet COVA2-05 competitively binds CR3022 but not EY6A. The researchers concluded, based on this pattern, that these five antibodies have overlapping yet different epitopes. Especially important is that CR3022 has, on its light chain, a CDR loop 1, which extends beyond the light chain of EY6A, and which may cause steric hindrance in the interaction between EY6A and NBP10 and NBP11.
Binding angles may thus be an important area of study in achieving neutralization. While CR3022 and EY6A strongly compete with each other for RBD binding, only the latter has been found to have neutralizing activity against SARS-CoV-2 entry, and this area requires further study.
ABP18 is an IGHV1-69 antibody. ABP11 also competes with the ACE2-binding antibodies like COVA1-12, COVA2-04, and COVA2-07, but not with others.
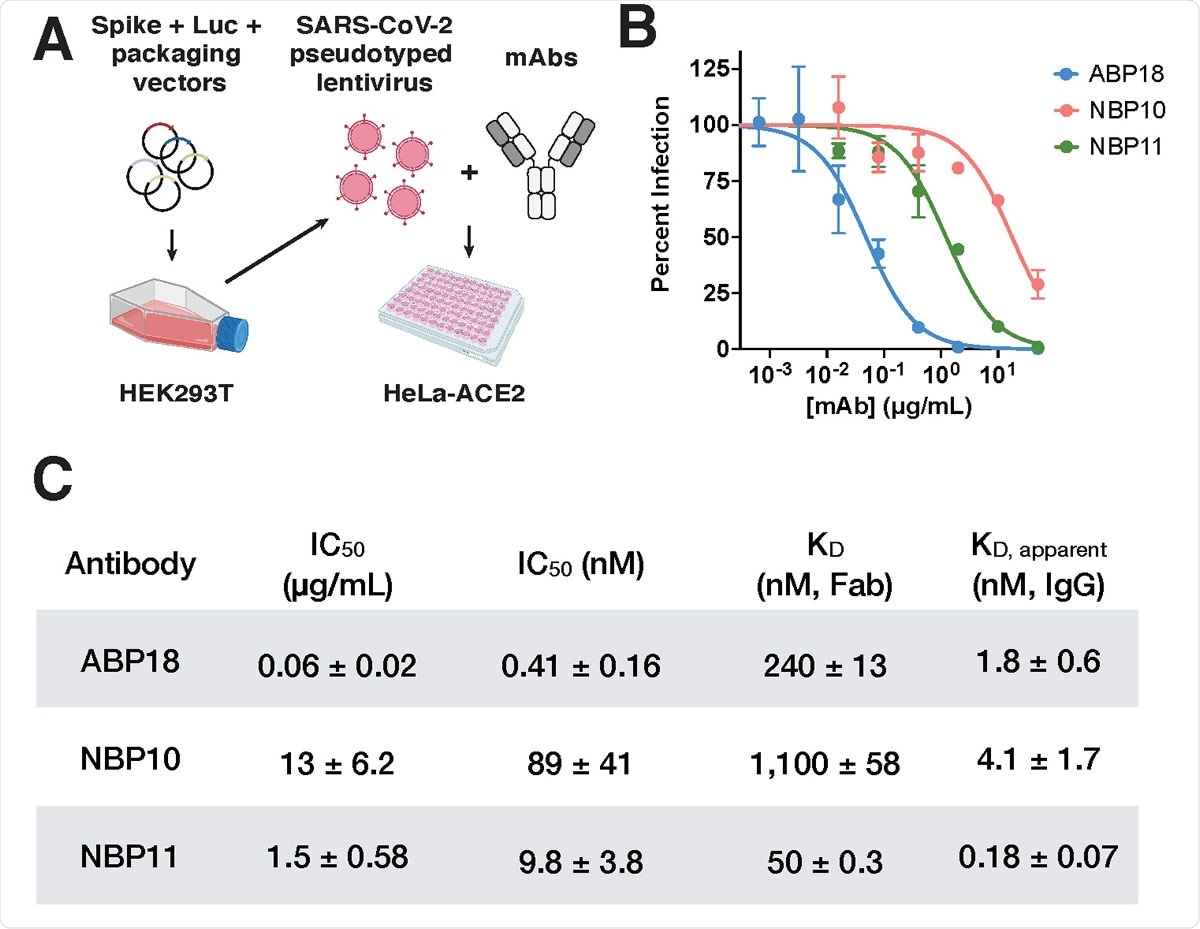
RBD antibodies neutralize SARS-CoV-2 pseudotyped lentivirus (A) SARS-CoV-2 pseudotyped lentivirus produced in HEK293T cells encode firefly luciferase (Luc) and can infect HeLa cells stably expressing the ACE2 receptor (HeLa- ACE2)19,20. Half maximal inhibitory concentration (IC50) can be determined by titrating a monoclonal antibody (mAb) of interest. Figure created with BioRender.com. (B-C) ABP, NBP10, and NBP11 IgG (B) neutralize SARS-CoV-2 spike-pseudotyped lentivirus in vitro with (C) IC50 values (mean ± standard deviation) consistent with their true or apparent affinities. Representative inhibition curves are shown from repeated (n=3) experiments.
What are the implications?
Using all available binding and competition data, along with already published structural data, the investigators concluded that the RBD has several unique epitopes and that these are targeted by separate monoclonal antibodies. Moreover, the presence of distinct footprints and competitive binding patterns even among ACE2-binding antibodies indicates that these monoclonal ACE2-targeting antibodies operate via a host of mechanisms to neutralize viral entry.
Antibodies with low somatic hypermutation are found to effectively neutralize the current virus. Moreover, even anti-RBD antibodies from the same germline may be directed against different epitopes.
“This observation is particularly promising for RBD- and spike-based vaccination strategies, as they may readily elicit neutralizing antibodies in patients presenting varied human antibody repertoires.”
The use of in vitro naïve antibodies selected to counter such binding could mirror the in vivo antibody response, helping to identify neutralizing antibodies against SARS-CoV-2 as well as other novel pathogens.
“Additionally, these novel RBD-targeting antibodies demonstrate the numerous distinct epitopes that can be effectively targeted on the SARS-CoV-2 RBD, bolstering COVID-19 therapeutic and vaccine efforts.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources