As the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to cause cases of coronavirus disease 2019 (COVID-19) throughout the world, early detection of cases is a high priority to minimize the transmission of the virus during the presymptomatic phase. An interesting new preprint research paper that was posted on the bioRxiv* server recently describes the use of rigid monoclonal antibodies (mAbs) in contrast to flexible mAbs, and how this affects the binding of multiple mAbs to the same antigen.
The SARS-CoV-2 nucleocapsid protein (NP) is essential for the incorporation of viral genomic RNA into new viral particles. Their synthesis occurs within the replication-transcription complexes formed within the host cell, the site of RNA replication. The N protein is produced in abundance and elicits a robust immune reaction. However, most assays target the spike antigen, citing lower cross-reactivity among antibodies and higher correlation with neutralizing capacity.
A successful NP assay could make diagnosis much more straightforward. Serological assays cannot be used before 7-10 days post-infection, this being the minimum period for seroconversion. Antigen-based detection is the most sensitive method for the diagnosis of SARS-CoV-2 infection. In addition, this type of test is well-suited to point-of-care lateral flow assays (LFA).
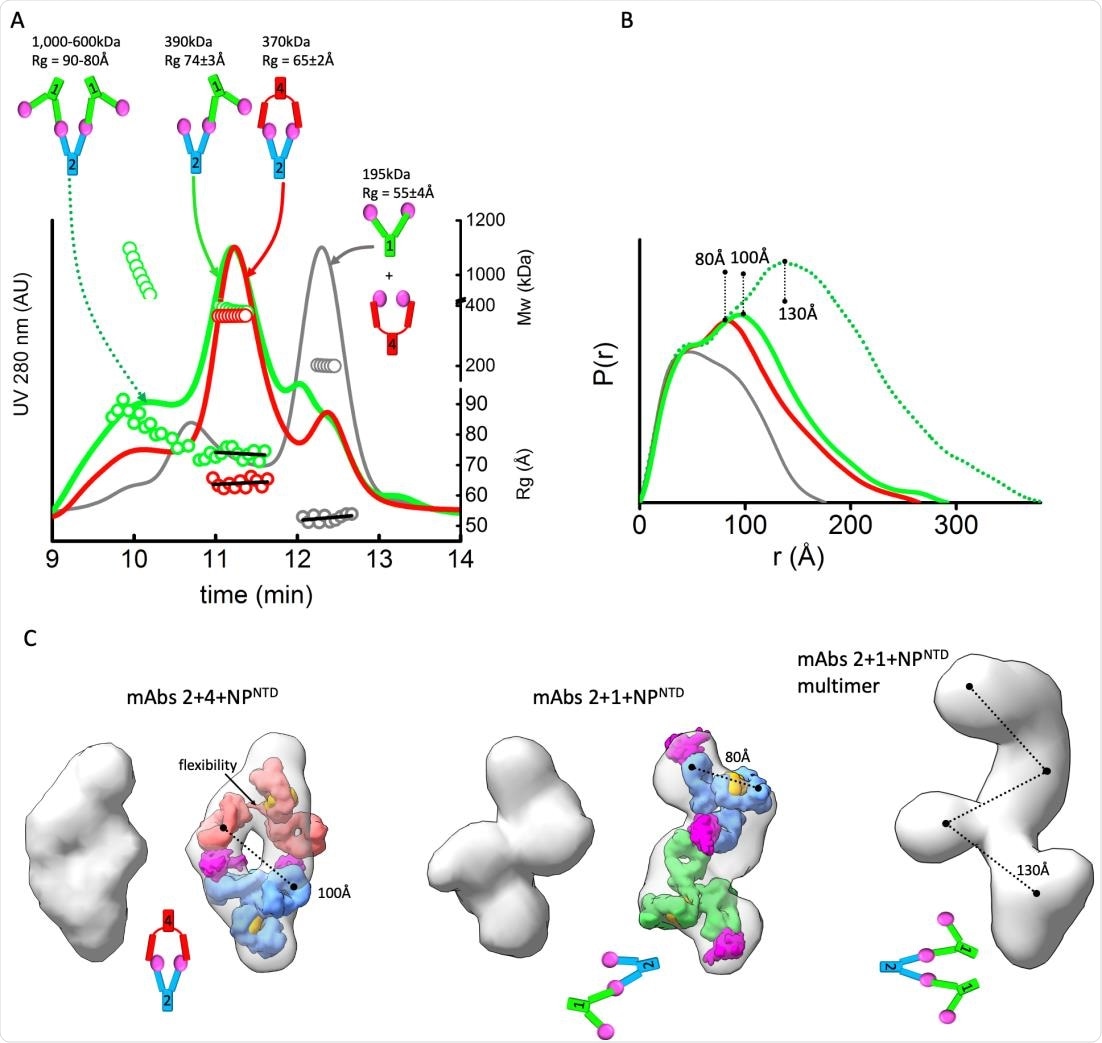
MAbs linear or sandwich pairing depends on inherent flexibility. A) SEC-MALS-SAXS chromatograms for the mAb1-2-NPNTD (green), mAb2-4-NPNTD (red) and mAb1-4-NPNTD (gray) samples. Solid lines represent the UV 280nm signal in arbitrary units, while symbols represent molecular mass (top) calculated from MALS and Rg values (bottom) for each collected SAXS frame versus elution time. B) P(r) functions calculated for the experimental SAXS curves for the main SEC peak of mAb1-2-NPNTD (green), mAb2-4-NPNTD (red), mAb1-4-NPNTD (gray), and early SEC shoulder of mAb1-2-NPNTD (green dots). The P(r) functions are normalized at the r=40Å. The P(r)-maxima peaks are indicated. Experimental SAXS and Guinier plots are shown in Supplemental Figure 1. C) Average SAXS envelope obtained for mAb2-4-NPNTD, mAb1-2-NPNTD complex were calculated using P2 symmetry operator.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Potential of LFAs
LFAs are not as sensitive as the reverse transcriptase-polymerase chain reaction, but this is an area that is being worked on actively. For these tests, the NP antigen has a distinct advantage in its abundance and structure. The NP is a phosphoprotein with two domains and an Arg-Ser linker. The protein exists as a dimer through the C-terminal domain (CTD).
In the presence of RNA, both the N-terminal domain and the CTD of the NP form a single interaction site. This forms the foundation on which the nucleocapsid is built. NP detection is possible for up to one day before symptom onset, and fluorescence LFA assays are currently available for the detection of the SARS-CoV-2 NP in both nasopharyngeal and nasal swab specimens.
The LFA test methods could exploit the agglutination of antigens, mediated by antibodies. The higher the antigen valency, the greater the antigen-antibody complex formation. Agglutination also occurs with paired mAbs that bind to separate epitopes on the antigen. LFAs based on this phenomenon has led to better sensitivity and specificity, lowering the limit of detection of these tests for NP.
Antibody rigidity and complex arrangement
Antibody flexibility is an important parameter in mAb recognition, and therefore in LFA performance. In the current study, the researchers used biophysical methods to characterize mAbs targeting the NTD of the NP from a pool of nine commercial anti-NP antibodies. This was used to correlate the antibody's flexibility with the superstructure formed following the binding of mAbs to the NTD of the NP.
The researchers used multiple techniques, including size exclusion chromatography (SEC) coupled with small-angle X-ray scattering (SAXS) and multi-angle light scattering (SEC-MALS-SAXS), to identify mAbs that bind the NP-NTD selectively. They coupled these findings with atomic models of mAbs in order to visualize the structure of the multi-antibody complexes, especially the conformation of the Fc to the Fab fragments.
Rigidity results in linear arrangement
They performed molecular dynamics (MD) simulations on the Fc-hinge region at extreme temperatures so that the heat keeps the Fabs from being trapped. They found it was simple to differentiate the pairs of mAbs that bind different epitopes of the NP-NTD from those which simply compete for the same binding site. They realized that mAb1 and mAb4 belong to the latter category, while mAb1 or mAb4 can be mixed with mAb2 to form species of higher masses, indicating they bind different epitopes.
They found a different stoichiometry and orientation for each mAb pair, as well as different masses. For instance, the 380 kDa mAb1-mAb2 complex has a different orientation between the pairs relative to the 340 kDa mAb2-mAb4 complex, with a linear vs. a sandwich-like arrangement that is hollow at the center. The ‘sandwich’ corresponds to the envelope and is made up of two antigens bound by two Fabs, one on each side. The linear mAb1-mAb2 pair shares a single NTD.
The researchers attribute this difference in orientation to variations in the flexibility of the mAbs. In the mAb2-mAb4 pair, both are flexible, resulting in the mAbs being closed and capped around both the antigens. The more rigid Fab of mAb2 is thus able to include two NTDs of two NPs, because of the highly flexible mAb4. Since mAb1 also has a rigid Fab, it cannot bind with the NP-NTD on the mAb2, causing the conformation of the complex to be linear.
Linearity allows polymerization
The linear arrangement of this complex allows for many mAbs to form a network through the epitopes left uncovered on the NTDs attached to the most external regions of the Fabs. The resulting complex can become very large, perhaps because the linear conformation is extended. Thus, agglutination is influenced by mAb flexibility.
The difference in functional outcome cannot be attributed to differences in affinity, because all antibodies' kinetics were found to be comparable at picomolar levels.
Higher signal from linear mAb complex
The researchers also found that the detection sensitivity for the linear mAb complex could be boosted by a modification of the conventional enzyme-linked immunosorbent assay (ELISA) protocol, with the detection mAbs being added to the test antigens during the incubation period and spiking free mAb2 into the detection mAbs prior to this addition. This was to allow maximal antibody polymerization, with the spiking later on leading to extended network formation of the linear arrangement. Such polymerization cannot occur with the sandwich arrangement. The LOD was lowered, accordingly, by two-fold, at high antigen concentrations.
This structurally-derived enhancement of sensitivity was sustained even when a detergent was added, thus disrupting the virion. In fact, the presence of the detergent Triton-X-100 boosted the LOD below 0.4 pg/mL.
What are the implications?
While the US FDA has recently approved an LFA over-the-counter test for at-home use in the detection of SARS-CoV-2N antigens, the LOD of such tests requires to be increased. Most such tests use colorimetric mAb-based assays, enhanced by luminescent nanoparticles, fluorescent immunoassays or magnetic beads. Unlike PCR, however, the signal is not amplified when the test antigen is detected by the probe.
Secondly, such assays use antigen-antibody binding at a ratio of 1:1, but increasing the ratio of detection antibodies to capture antibodies will increase the ratio of the signal to the antigen as well, and thus enhance its detection. This is where the current study helps by providing a rapid method to first identify mAb pairs that bind linearly to the antigen, which increases detection sensitivity. This approach is easy to use and allows antibody interactions to be studied in solutions, where they are more dynamic relative to studying them in crystals or in grids.
The observers also found that the flexibility of mAbs can affect larger assemblies of mAbs even without additional antigens. The tendency of many mAbs to aggregate has prematurely ended many development processes. However, the current approach can rapidly separate such aggregates from single mAbs, thus relating structure with aggregation. It can also differentiate flexible and rigid mAbs in solution. More research is required to understand how far glycosylation affects flexibility.
The polymerization of the mAb-mAb4 seen here would be dismissed as aggregation in other techniques but is here exploited to amplify the signal following antigen detection in an antigen concentration-dependent manner.
Further optimization is possible, without adding external factors, by making use of the inherent rigidity of the mAbs. Using this approach, hundreds of mAbs can be screened to find the most rigid, and then the LFA LOD reduced by such optimization methods to develop a point-of-care device with high sensitivity for detection of SARS-CoV-2 as well as other future pathogens.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources