The emergence of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants has been of grave concern to public health authorities and governments involved in stopping the ongoing pandemic of coronavirus disease (COVID-19). A new preprint research paper posted to the medRxiv* server deals with the preliminary results of a study on the so-called British variant, UI-202012/01, also known as lineage B.1.1.7 or 20B/501Y.V1.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This variant was first documented on December 14, 2021, and is defined by the N501Y mutation in the receptor-binding domain (RBD) of the spike protein, the deletion ΔH69/V70, and many other mutations. It has recently been reported that this variant is rising in frequency in the South-east of England so fast as to raise the suspicion that it has increased transmissibility.
The current study aimed to investigate this aspect of the virus in terms of increased viral load. The researchers sequenced all positive samples from four Lighthouse laboratories in the UK, using their quantitative sequencing approach. This yields the number of unique mapped reads, which bears a correlation with and therefore acts as a proxy for the viral load.
They found that the logarithm of unique mapped reads was negatively correlated with the Ct values obtained from the polymerase chain reaction (PCR) testing. They selected the presence of Y501 as a marker of the new variant. From 88 samples that showed this mutation, they considered only the samples taken in the period from October 31 to November 13, 2020. This showed that the number of unique mapped reads in the Y501 variant was more significant than in the N501 variant, indicating that the median viral loads are increased by about three times for the Y501 variant.
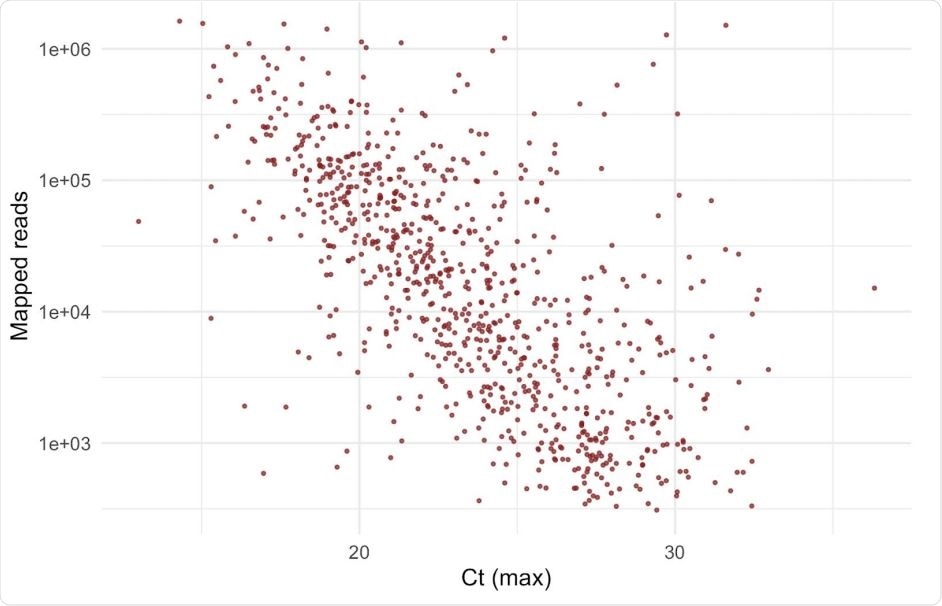
A strong negative correlation between Ct value and log10(number of mapped reads). Number of uniquely mapped reads per sample can be used as a proxy for viral load. The Ct value shown is the maximum Ct value obtained from Majora (the COG database) from retrospective data for all Lighthouse laboratories that supply Ct data; log10 of uniquely mapped (deduplicated) reads obtained with veSEQ platform correlates well with Ct. This does not include samples in this report since Ct values were not yet available.
Viral loads differ by location
The researchers then examined the geographical location from which different samples were taken, where such information was available. They found that whichever variant was present, all samples from Greater London had a much higher viral load compared to any other location.
At other locations, however, the viral loads for Y501 were much higher compared to N501. Significantly, however, the location, which was enriched for Y501, Kent county, did not show any difference between Y501 and N501 with respect to viral loads. This could indicate that the infection with VUI-202012/01 and the viral load were correlated outside Greater London. In the latter location, the reasons for the lack of association could be because the study was underpowered, or alternatively, it could be due to differences in the demographic and epidemiologic parameters of the London population relative to other locations.
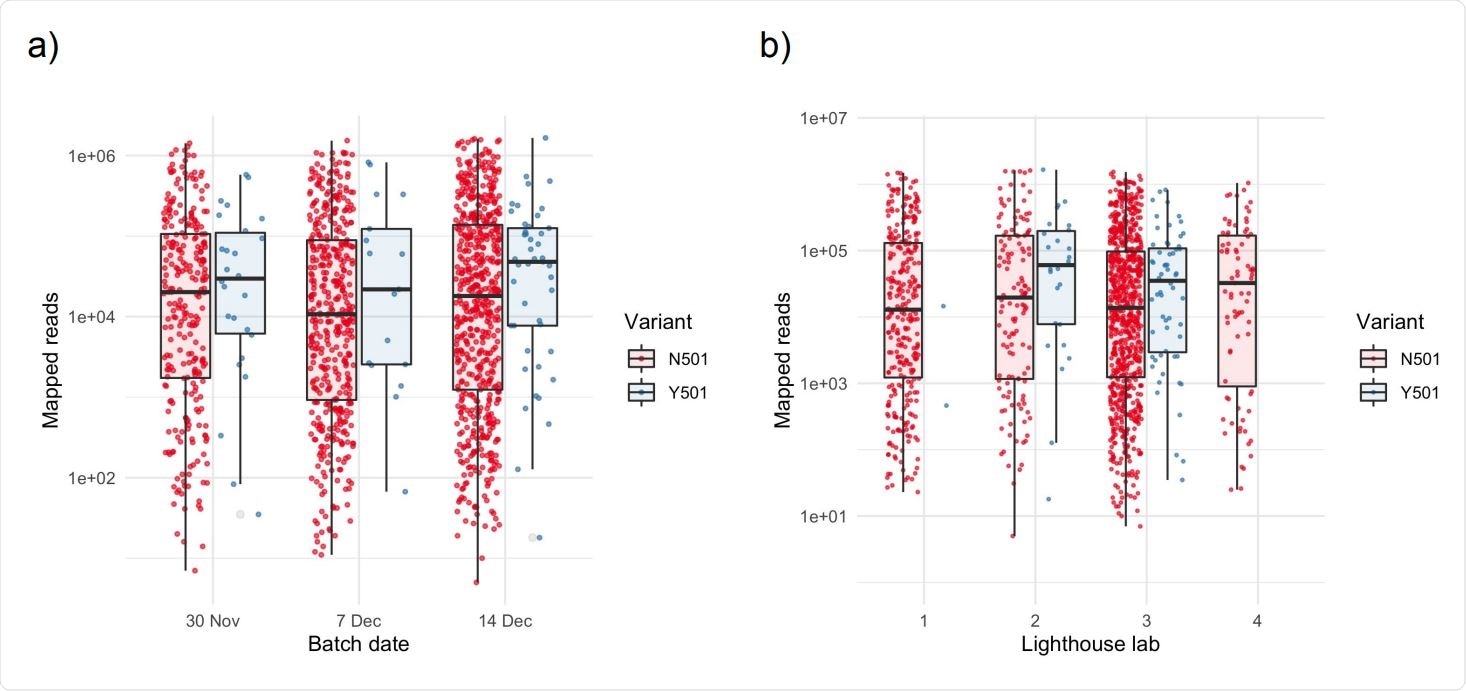
Higher numbers of mapped reads in samples exhibiting the Y501 variant Box and scatter plots of unique mapped reads, stratified by (a) batch date and (b) anonymized Lighthouse lab. There is no significant difference among batches or Lighthouse labs for N501 samples (p>0.1 for all pairwise comparisons). Points within each batch are jittered to aid visualization. Horizontal lines in boxplots represent the median and the interquartile range.
What are the implications?
The researchers point out that the samples in this study came from people who were symptomatic. The viral loads are thought to be higher at the start of symptoms. Since the sequenced data covered a broad range of viral loads, they suggest that the test data came from individuals at all stages of symptomatic infection.
The new variant may be circulating in a particular age group, for instance, which has higher viral loads. This may be one explanation for the difference in Ct-viral load correlations in Greater London vs. outside this region. Another factor is the intense transmission that occurs during the initial phase of an epidemic caused by a given lineage. Dispersion of the lineage occurs much later. This can be confirmed only with more research.
The reasons for the faster growth associated with this variant are not clear: it could be due to faster epidemic growth, demographic patterns, founder effects, or higher viral loads, among other biological mechanisms. The correlation with higher viral loads, in this case, seems to suggest increased transmissibility of this virus, but further studies are required. Again, the N501Y mutation may not be the only reason for this expansion. There is more need to understand how viral levels are related to virulence since this may determine the infection's severity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources