The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused thousands upon thousands of infections the world over, in the ongoing pandemic of coronavirus disease 2019 (COVID-19). It is vital to understand the structure of this virus's genome to develop new antiviral therapies.
The SARS-CoV-2 is a positive-sense RNA virus with a genome that has approximately 30 kilobases. Its RNA-dependent RNA synthesis comprises two distinct processes, namely, making copies of the genomic RNA (gRNA) by continuous replication, and transcribing copies of the genes encoding the various viral structural and accessory protein by discontinuous transcription of subgenomic RNA (sgRNA). Transcription regulating sequences (TRS) at the 3' end of the leader sequence as well as proximal to each gene, called TRS-L and TRS-B, respectively, join to the body via the base pairing of the core sequences (CS) of the leader TRS and the complementary strand core sequence (cCS) for each body or viral gene.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The importance of high-order structures
Earlier studies have suggested that RNA-RNA interactions over long distances are essential to template switching. For instance, the B motif and its complementary motif must interact to allow higher-order structures to form, such that sgRNA can be formed. However, this knowledge comes from other coronavirus studies, and the motifs are not conserved. The researchers, therefore, assume rather than know that the TRS-L interacts with the cCS-B, and also with TRS-B.
In the current study, the scientists compared the TRS-B chimeric reads that underwent ligation in two opposite orientations, discovering two classes of reads. One was the result of RNA-RNA interactions and the other represented sgRNAs.
Sanger sequencing was carried out as a validation test, confirming two of the putative sgRNAs. It was found that despite the lack of canonical cCS-B motifs preceding the body of the gene in either of these novel sgRNAs, there was a partial overlap of both sgRNAs with the canonical CS motif. In other words, base pairing at the TRS-L and, at least in part, at the cCS-B, are essential for template-switching while discontinuous replication is going on for these sgRNAs.
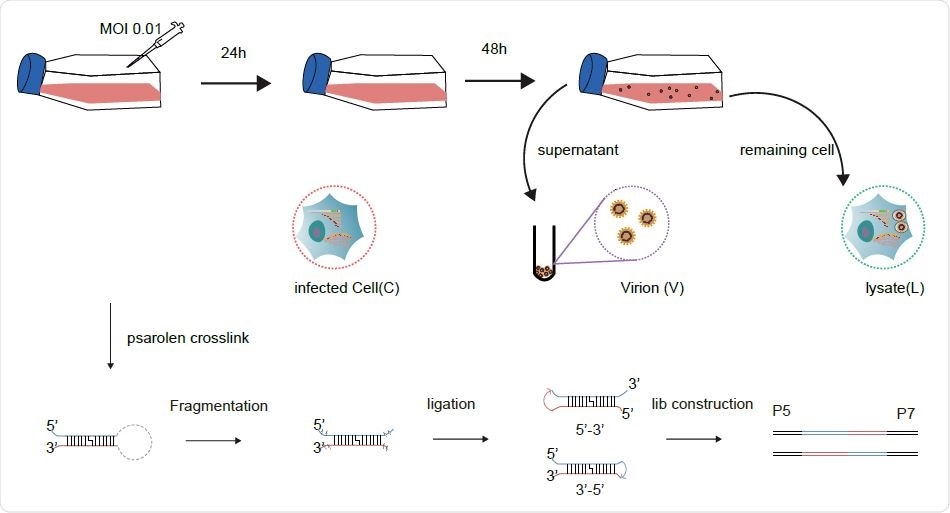
Schematic diagram for sample collection and major experimental steps.
Frameshifting element
The FSE is vital in the process of translating the nonstructural proteins in the viral open reading frame ORF1b, and any disturbance at this point could disrupt replication. Many earlier studies have shown the FSE in SARS-CoV-2 to comprise a pseudoknot of three stems, composed of mRNA, but higher-order structures as long as 1.5 kb have been shown to bridge the 3' and 5' ends of the ORF1a and the ORF1b, respectively. This so-called FSE arch is here suggested to be one of several alternative structures. The researchers propose a dynamic change in the regions around the FSE, with changing RNA conformations to fit the need of the moment, as regards the rate of frameshifting or the stoichiometry of nonstructural viral proteins.
Such non-canonical structures may form a larger high-order pseudoknot, along with Ziv's arch, which incorporates the FSE. Both large and small pseudoknots, thus formed, can explain the structure that causes ribosome stalling. More research is required to understand how these structures are chosen, all the more since these complex conformations are found even within the newly assembled viral particles.
Domain-specific structure
The genome was found to consist of several domains, and each domain shows strong interactions within itself, relative to extra-domain interactions. The domains are uniformly and regularly folded, rather like nucleosomes in eukaryotic genomes. Over the lifecycle of the virus, the domain boundaries remain constant.
Domain structure becomes still more prominent in virions, indicating that these flexible domain boundaries may present therapeutic sites for drug design.
As previously observed in the Zika virus, such domain-dependent genomic structure might be the standard in all single-stranded RNA viruses and hypothesize the researchers. In this case, the nucleocapsid (N) protein of the virus that packages RNA into new virions may be key to regulating genome structure once the virus enters a host cell and releases its RNA.
What are the implications?
The use of a simplified SPLASH protocol allowed for a higher RNA yield, sufficient to build a library and carry out deep sequencing studies. The use of RNase III allowed RNA fragmentation to occur, allowing for more efficient RNA ligation of all the treated RNA. The result was that they obtained ~30% chimeric reads in virions and over 10% chimeras in cells and lysates, the former probably being due to the increased compactness of the genome.
This study proves directly for the first time the presence of long-distance interactions between TRS-L and TRS-B regions. RNA secondary structures mediate the replication, transcription and translation of viral RNA and proteins for successful infection of the host cell.
The study also highlights the compaction of the SARS-CoV2 genome within the virion vs. within the host cell, accompanied by lower degrees of cyclization and interactions mediated by sgRNA in the former setting. Specific TRS-L interactions are more prominent in the virions.
The delineation of such structural elements underlying transcription regulation and other key viral replication processes should promote the development of better antiviral strategies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources