Researchers in South Africa have conducted a study showing that the novel 501Y.V2 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has emerged in the country is able to escape the neutralizing antibodies that are elicited by previously circulating strains of the virus.
SARS-CoV-2 is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that is currently sweeping the globe, devastating public health and the global economy.
The study found that the 501Y.V2 lineage also conferred complete escape from three classes of therapeutic monoclonal antibodies.
Penny Moore from the National Health Laboratory Service (NHLS) in Johannesburg and colleagues say the findings highlight the possibility of re-infection among people presumed to have acquired some degree of immunity due to previously having had SARS-CoV-2.
The findings also have important implications regarding the effectiveness of certain vaccines and therapeutic strategies that are undergoing development.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Two immunodominant spike regions are usually targeted by neutralization
The novel 501Y.V2 lineage contains multiple mutations within two immunodominant regions of the viral spike protein – the main structure SARS-CoV-2 uses to bind to and infect host cells.
One of these regions – the receptor-binding domain (RBD) – is the primary target of antibody neutralization during infection.
“Neutralizing antibodies are considered the primary correlate of protection against re-infection for most vaccines and are being actively pursued as therapeutics,” writes the team. “The overwhelming majority of monoclonal neutralizing antibodies isolated thus far from infected individuals, immunoglobulin libraries, or immunized animal models, target this region.”
The next most immunodominant region is the N-terminal domain (NTD) of the spike protein.
This domain is also frequently targeted by neutralizing antibodies and several potent monoclonal antibodies that target this region are currently being considered for clinical development, says Moore and colleagues.
The 501Y.V2 lineage has nine mutations across these regions
The 501Y.V2 lineage of SARS-CoV-2 that has recently emerged in South Africa contains nine mutations in the spike protein. One cluster is seen in the NTD and includes four substitutions and one deletion (L18F, D80A, D215G, Δ242-244, and R246I). Another cluster is found in the RBD and consists of three substitutions (K417N, E484K, and N501Y).
“Although the 501Y change has been associated with increased transmissibility, rather than immune pressure, the accumulation of mutations specifically within these two immunodominant regions of spike is highly suggestive of escape from neutralization,” write the researchers. “Indeed, mutations at 484 have been shown to reduce neutralization sensitivity,” they add.
In addition, mutations in the RBD and NTD of spike have also been described in a novel variant that has recently emerged in Brazil. Furthermore, variants in the UK and United States have also been described, suggesting that new SARS-CoV-2 variants are emerging globally.
What did the researchers do?
The team showed that spike mutations in the 501Y.V2 lineage exhibited complete neutralization escape from three classes of SARS-CoV-2-directed monoclonal antibodies.
Furthermore, this variant showed substantial or complete escape from neutralizing antibodies in plasma taken from individuals who had recovered from COVID-19.
Importantly, the team showed that the K417N mutation played a crucial role in the viral escape. This mutation effectively abrogated neutralization by a well-defined, multi-donor class of antibodies that make up some of the most common and potent neutralizing antibodies to SARS-CoV-2.
“Crucially, it is from these same public antibody responses that many therapeutic strategies currently under development have been derived,” writes Moore and the team. “The overwhelming majority of monoclonal antibodies already on the path to licensure target residues K417 or E484 and will therefore be ineffective against 501Y.V2.”
The researchers also defined an important role for a small three-amino-acid deletion in a relatively large region in NTD that completely disrupted a dominant antibody response to a site called the N5-loop or “supersite loop.”
“This deletion predominates among 501Y.V2 variants and occurs either alone or with an R246I substitution that has also been shown to abrogate neutralization by several NTD-directed neutralizing antibodies,” says the team.
The researchers also point out that a next generation of potent neutralizing antibodies targeting this NTD N5-loop supersite has been suggested for clinical development, but these are unlikely to be effective against 501Y.V2.
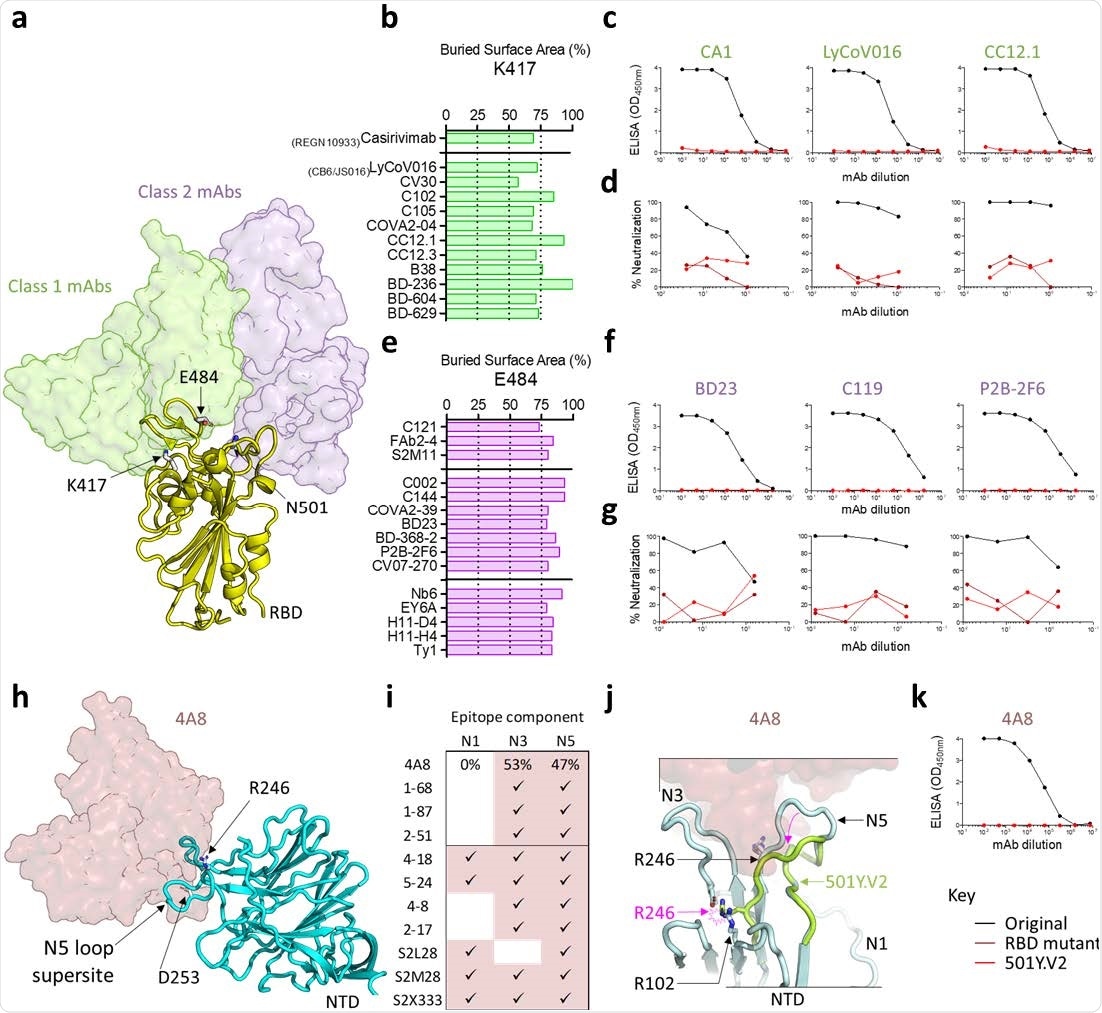
SARS-CoV-2 501Y.V2 is resistant to monoclonal antibodies to both RBD and NTD a. A structure of the SARS-CoV-2 RBD is shown in yellow cartoon view, modeled in complex with a representative neutralizing antibody from class 1 (CV30) and class 2 (C104). The VH domains of the representative class 1 antibody are shown in translucent green surface view, while those of the class 2 antibody are shown in translucent purple. The side chains of residues K417, E484, and N501 are indicated with arrows. b. A plot showing the percentage of accessible surface area for the RBD K417 side chain (x-axis) that is buried in the paratopes of several class 1 neutralizing antibodies (listed on the y-axis). VH3-53/66 antibodies are separated below the horizontal line. c. ELISA binding curves for antibodies CA1, LyCoV016, and CC12.1 to the original RBD (black) or the 501Y.V2 RBD (red). d. Neutralization curves for the same antibodies shown in c, against the original virus (black), 501Y.V2 (red), or a chimeric construct that includes only the RBD mutations K417N, E484K, and N501Y (maroon). e. Percentage of accessible surface area for the RBD E484 side chain (x-axis) that is buried within the paratopes of several class 2 neutralizing antibodies (listed on the y-axis). VH1-2 (top) or VH-diverse (middle) antibodies and nanobodies (bottom) are separated with the horizontal lines. f. ELISA binding curves for antibodies BD23, C119, and P2B-2F6 to the original RBD (black) or the 501Y.V2 RBD (red). g. Neutralization curves for the same antibodies shown in (f), against the original virus (black), 501Y.V2 (red), or the chimeric RBD construct (maroon). h. A structure of the SARS-CoV-2 NTD is shown in cyan cartoon view, modeled in complex with a VH1-24 neutralizing antibody 4A8 (shown in translucent maroon). The N5-loop supersite and side chains of residues R246 and D253 are indicated with arrows. i. The relative percent contribution of NTD loops N1, N3, and N5 to the 4A8 epitope were calculated, and contributions for additional NTD-directed neutralizing antibodies are shown. j. Modelling of the 501Y.V2 Δ242-244 deletion (lime green cartoon view), in the context of the 4A8 epitope. NTD loops N1, N3, and N5 are shown and the position of R246 in the original NTD or 501Y.V2 NTD is labeled with black or pink, respectively. The minimum displacement for 501Y.V2 loop N5, and the accompanying clash with R102 are indicated with the pink arrows. k. ELISA binding curves for 4A8 to the original NTD (black) or the 501Y.V2 NTD (red).
What do the researchers conclude?
The team says the findings suggest that while many people worldwide have already been infected with SARS-CoV-2 globally and are presumed to have accumulated some level of immunity, new variants such as 501Y.V2 pose a significant risk of re-infection.
The findings also raise important questions about the effectiveness of current spike-based therapeutic strategies and vaccines.
“Altogether, these data highlight the need for increased, ongoing surveillance and sequencing during the SARS-CoV-2 pandemic,” writes Moore and colleagues.
“The speed and scope of 501Y.V2 mediated immune escape from pre-existing neutralizing antibodies highlight the urgent requirement for rapidly adaptable vaccine design platforms, and the need to identify less mutable viral targets for incorporation into future immunogens,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Moore P, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv, 2020. doi: https://doi.org/10.1101/2021.01.18.427166, https://www.biorxiv.org/content/10.1101/2021.01.18.427166v1
- Peer reviewed and published scientific report.
Wibmer, Constantinos Kurt, Frances Ayres, Tandile Hermanus, Mashudu Madzivhandila, Prudence Kgagudi, Brent Oosthuysen, Bronwen E. Lambson, et al. 2021. “SARS-CoV-2 501Y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma.” Nature Medicine 27 (4): 622–25. https://doi.org/10.1038/s41591-021-01285-x. https://www.nature.com/articles/s41591-021-01285-x.