Researchers at the University of Michigan in the United States have cautioned against only using within-host minor genetic variations to resolve the genomic epidemiology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
Adam Lauring and colleagues found that intrahost single nucleotide variants (iSNVs) can arise in SARS-CoV-2 genomes that are not related through local transmission. The variants are not necessarily predictive of subsequent global spread of those mutations.
The team says the findings suggest that intrahost variants may have be of limited utility for predicting future lineages.
“These results provide important context for sequence-based inference in SARS-CoV-2 evolution and epidemiology,” warn the researchers. “Because iSNVs can arise in parallel in genomes that are not linked by transmission, caution is needed when relying entirely on shared iSNVs for transmission inference.”
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
Study: Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. Image Credit: Orpheus FX / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
How do patterns of intrahost diversity help?
Genomic epidemiology is necessarily constrained in its ability to determine the exact transmission chains that arise during an outbreak, such as COVID-19.
Patterns of shared intrahost variants between individuals infected with a virus can indicate the relative importance of natural selection, reveal genetic loci under convergent evolution, and enable measurement of the transmission bottleneck, which is critical in determining the spread of new variants.
However, while a clear understanding of within-host evolution could help to inform how SARS-CoV-2 spreads on a broad scale, few studies have conducted comprehensive analyses of intrahost dynamics.
“Little is known about temporal aspects of SARS-CoV-2 intrahost diversity and the extent to which shared diversity reflects convergent evolution as opposed to transmission linkage,” writes the team.
Although early reports demonstrated that SARS-CoV-2 exhibits intrahost diversity, this has not been as well researched as consensus-level genomic diversity.
“Intrahost diversity is an important complement to consensus sequencing,” says Lauring and colleagues.
The researchers say that to determine the utility of SARS-CoV-2 intrahost diversity for transmission inference, an improved understanding of its temporal variation during infection and its convergent evolution across individuals is needed.
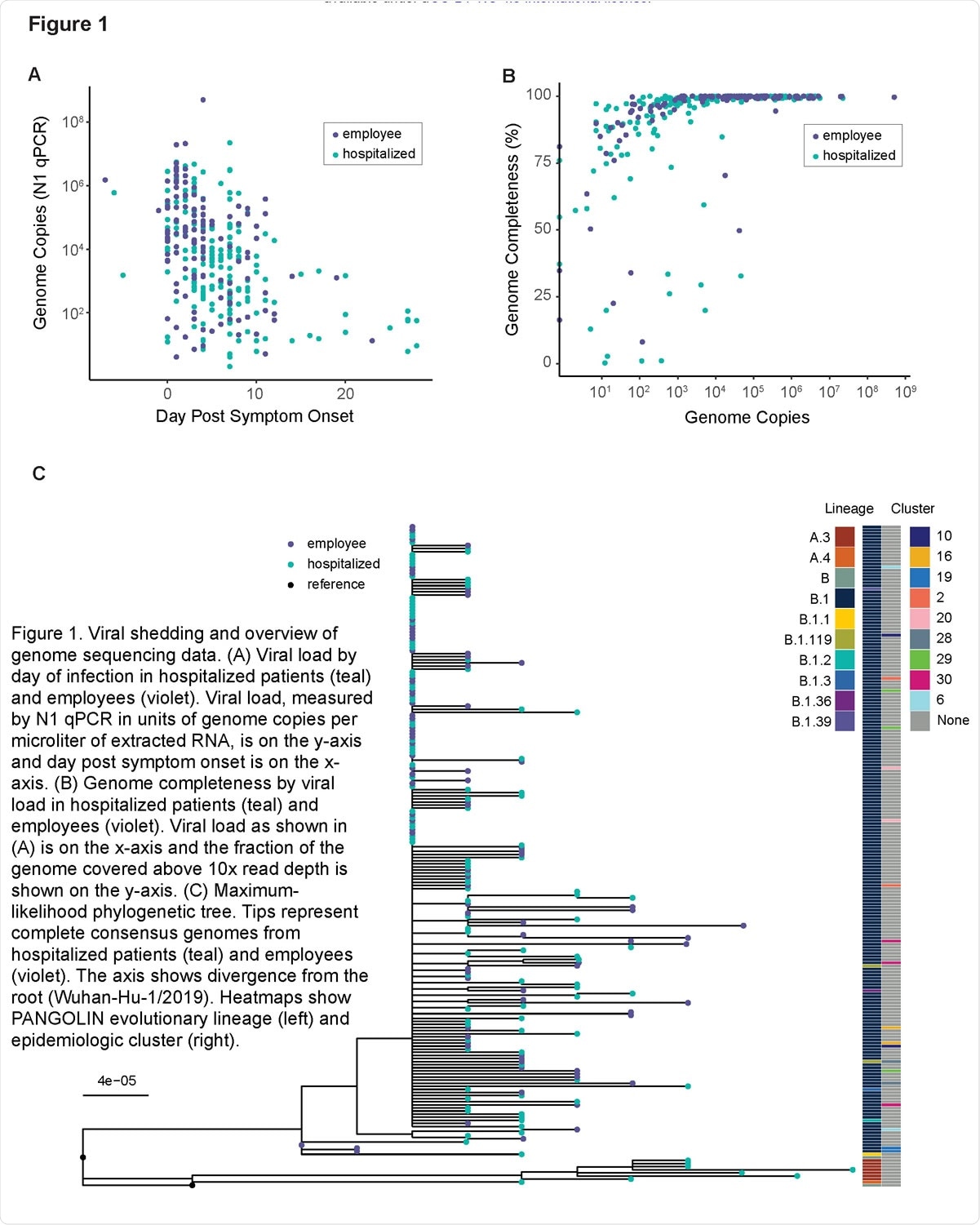
Viral shedding and overview of genome sequencing data. (A) Viral load by day of infection in hospitalized patients (teal) and employees (violet). Viral load, measured by N1 qPCR in units of genome copies per microliter of extracted RNA, is on the y-axis and day post symptom onset is on the x-axis. (B) Genome completeness by viral load in hospitalized patients (teal) and employees (violet). Viral load as shown in (A) is on the x-axis and the fraction of the genome covered above 10x read depth is shown on the y-axis. (C) Maximum-likelihood phylogenetic tree. Tips represent complete consensus genomes from hospitalized patients (teal) and employees (violet). The axis shows divergence from the root (Wuhan-Hu-1/2019). Heatmaps show PANGOLIN evolutionary lineage (left) and epidemiologic cluster (right).
What did the researchers do?
The team performed high depth-of-coverage sequencing of 212 complete SARS-CoV-2 genomes obtained from 325 respiratory specimens. The samples were collected between March and May 2020 from 65 hospitalized COVID-19 patients (190 samples) and infected employees (135 samples) at a single medical center.
The team found that intrahost diversity is low and its distribution does not significantly vary by time since symptom onset.
Since the identification of viral intrahost variants can be error-prone, the team sequenced defined RNA mixtures to validate the accuracy of their variant calling pipeline.
This revealed that low input viral load decreases the specificity of variant calling and that sufficient input is a critical factor for accurate identification of iSNV.
“Future studies of SARS-CoV-2 intrahost diversity should report and account for specimen viral loads to avoid this common source of error,” advises the team.
What else did the study reveal?
Generally, there was little shared intrahost diversity across the cohort; most iSNVs were unique to each individual. However, 19 of the variants were identified in multiple specimens. Two variants (G12331A and A11782G) were present in three people, and one (U13914G) was present in six people.
There was no clear evidence of phylogenetic clustering of the genomes that shared the mutations.
The U13914G mutation was shared between multiple sample pairs separated by two or more substitutions, and G12331A was shared between samples from different viral lineages.
None of the three mutations (which were first detected in late March 2020) reached more than 1% frequency per week in consensus sequences submitted to GISAID (Global Initiative on Sharing Avian Influenza Data) through mid-November 2020.
“These results suggest that iSNV that arise convergently across viral lineages are not necessarily predictive of the subsequent global spread of those mutations,” writes the team.
Shared variants unlikely to be transmission-linked
Transmission inference that is based on shared intrahost variants incorporates data such as consensus genome sequences, date of sample collection, and shared iSNVs.
The researchers, therefore, compared shared iSNVs across all unique pairs of specimens used for variant calling.
The researchers identified 14 unique pairs, with shared iSNVs between genomes that were almost identical (0 to 1 consensus differences), eight of which were collected within one week of each other. However, there was no epidemiologic data to suggest that these pairs were linked by transmission.
The team also identified shared iSNVs between 23 pairs that were separated by two or more consensus substitutions and between 15 pairs with collection dates that were 7 to 28 days apart.
“Due to differences in viral lineage and time of collection, these are very unlikely to be transmission pairs,” say the researchers.
What does the team advise?
Lauring and colleagues say that since intrahost variants may be shared through parallel mutation rather than transmission, caution is warranted in the use of shared iSNV alone for inferring transmission chains.
“Unified statistical frameworks that incorporate sequences, metadata, and epidemiological models are likely the most robust approaches for integrating intrahost variants, but these models also must account for parallel evolution,” they advise.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Adam Lauring. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.01.19.427330, https://www.biorxiv.org/content/10.1101/2021.01.19.427330v1
- Peer reviewed and published scientific report.
Valesano, Andrew L., Kalee E. Rumfelt, Derek E. Dimcheff, Christopher N. Blair, William J. Fitzsimmons, Joshua G. Petrie, Emily T. Martin, and Adam S. Lauring. 2021. “Temporal Dynamics of SARS-CoV-2 Mutation Accumulation within and across Infected Hosts.” Edited by Andrew Pekosz. PLOS Pathogens 17 (4): e1009499. https://doi.org/10.1371/journal.ppat.1009499. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009499.