A team of international scientists has recently developed a bispecific antibody that can bind two non-overlapping regions of the spike receptor-binding domain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and completely prevent the viral-host interaction. The antibody is also capable of neutralizing different mutated variants of SARS-CoV-2. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Many studies have been undertaken to search for effective therapeutic interventions to curb the ever-growing trajectory of coronavirus disease 2019 (COVID-19). In this context, various pre-clinical and clinical studies have shown that monoclonal antibodies developed specifically against SARS-CoV-2 effectively treat COVID-19 patients. Moreover, it has been observed clinically that compared to a single antibody, a combination of two or more antibodies exhibits higher efficacy and increased potency in preventing viral escape. However, this approach is not cost-effective because of the complex and time-consuming manufacturing process.
In the current study, the scientists have conducted structural analysis and computational simulations to design a bispecific immunoglobulin G (IgG)-like molecule that can simultaneously bind to two distinct, non-overlapping epitopes on the same receptor binding domain (RBD) of SARS-CoV-2 spike protein, as well as different RBDs on a spike trimer.
Study design
Using a molecular dynamics simulation approach, they have designed four molecules, of which, CoV-X2 has shown the highest neutralizing efficacy against SARS-CoV-2 pseudoviruses. Specifically, CoV-X2, an IgG1-like bispecific antibody, has been developed by combining the Fragment antigen-binding (Fab) domain of two human monoclonal antibodies, namely C121 and C135, which are known to have significant SARS-CoV-2 neutralizing efficacy.
According to the structural predictions, CoV-X2 can bind to RBD at all conformations, spike trimer, and several spike trimer mutants, including the D614G-containing variant and the novel UK and South African variants of SARS-CoV-2. Moreover, CoV-X2 can bind to SARS-CoV-2 variants that cannot be neutralized by parental monoclonal antibodies (C121 and C135).
Surface Plasmon Resonance-based avidity assay has revealed that CoV-X2 can simultaneously bind to two distinct regions on the same RBD. The strength of the binding is maintained even at a low antigen concentration. Moreover, CoV-X2 has been found to completely inhibit the interaction between viral spike protein and host angiotensin-converting enzyme 2 (ACE2). In contrast, only partial inhibition of spike/ACE2 interaction has been achieved by both parental antibodies.
The neutralizing ability of CoV-X2 has been investigated using SARS-CoV-2 pseudoviruses. The findings have revealed that CoV-X2 is effective in neutralizing both wild-type and mutated variants of SARS-CoV-2. However, both parental antibodies have failed to neutralize mutation-bearing SARS-CoV-2.
The clinical efficacy of CoV-X2 has been determined by infecting human ACE2-expressing mice with SARS-CoV-2 and subsequently treating them with the antibody. The infected mice without CoV-X2 treatment have developed lung pathologies similar to severe COVID-19 patients, in addition to significant bodyweight loss. The viral RNA has been detected in the lungs and distant organs of these infected mice. In contrast, no significant loss of body weight and no histopathological alterations have been observed in CoV-X2-treated mice. Moreover, the CoV-X2 treatment has been found to significantly reduce the viral load as well as the frequency of detecting infectious virus, indicating the effectiveness of CoV-X2 in reducing the infection and disease.
To compare the anti-SARS-CoV-2 effectiveness of CoV-X2 and parental antibodies, the scientists have treated the virus-infected mice with individual parental antibodies. Interestingly, C121-treated mice have been found to acquire E484D mutation. The same mutation, which is pathogenic and known to reduce the virus neutralization efficacy of human antibodies by 10-fold, has also been observed in the novel South African variant of SARS-CoV-2. In contrast, none of the mice treated with CoV-X2 have developed pathogenic mutations, indicating the effectiveness of CoV-X2 in suppressing viral escape.
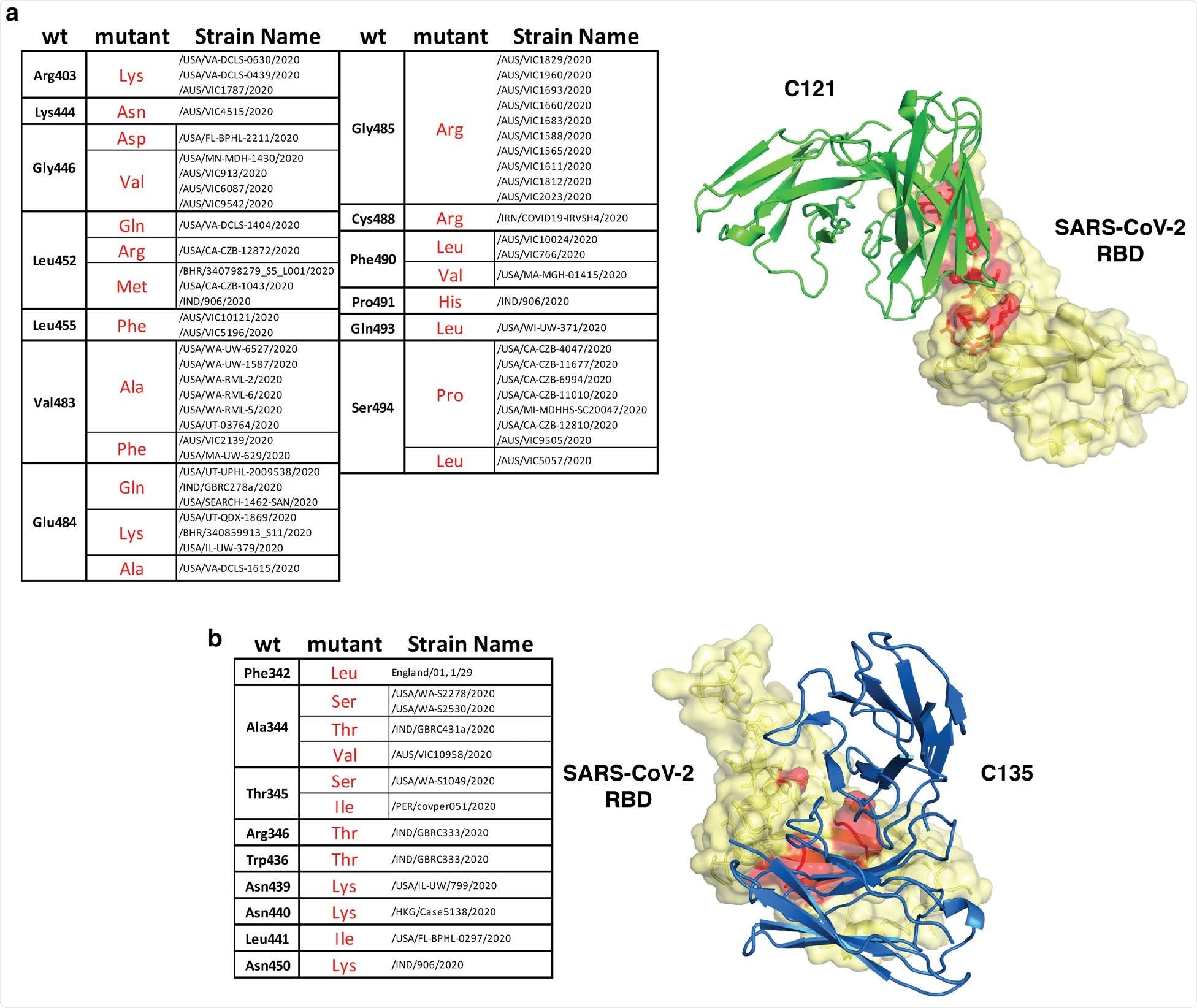
Natural SARS-CoV-2 variants in the C121 and C135 epitopes. Summary of naturally occurring mutations in the C121 (a) or C135 (b) epitopes reported in circulating SARS-CoV-2 (as of January 1, 2021). The location of the mutated residues is shown in red on the RBD structure. C121 and C135 variable regions are in green and blue (PDB ID: 7K8X and 7K8Z respectively).
Study significance
A bispecific IgG1-like antibody, CoV-X2, has been developed in the study that shows significant neutralizing efficacy against SARS-CoV-2. The antibody is capable of completely inhibiting the spike-ACE2 interaction and preventing the viral escape. Unlike parental antibodies (C121 and C135), CoV-X2 can effectively neutralize a wide-range of mutated variants of SARS-CoV-2, which makes it a potential candidate to combat the COVID-19 pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources