The UK and South African variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been suggested to have higher infectivity than the ancestral strain. The question remains, are they also more dangerous? Do they escape neutralization by antibodies elicited by the currently available vaccines?
A new preprint research paper by scientists from Sweden and India attempts to provide a preliminary answer to these questions using computational modeling. The research, which appears on the bioRxiv* server, predicts increased virulence with the new variant. If validated, this could help public health authorities prepare for new variants' possible impact as they emerge.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The UK variant
The new variant called Variant of Concern 202012/01, abbreviated VOC-202012/01, the year 2020, month 12, variant 01) belonging to the lineage B.1.1.7 was first sequenced in the United Kingdom. It has some strange multiple mutations in the spike protein, namely, N501Y and A570D in the receptor-binding domain (RBD) and the N terminal domain, respectively, H69/V70, 144/145 deletions in the S1 subunit besides P681H, T716I, S982A, and D1118H mutations in the S2 subunit.
Altogether, the UK variant has 29 mutations relative to the original Wuhan strain, representing an accelerated mutation rate compared to the predicted two mutations per month. Moreover, there is also an unexpectedly high number of non-synonymous mutations in the spike protein, with 27% of them being common to the ancestor. Many of these affect essential processes such as spike cleavage or change the sequence of a stop codon (the open reading frame 8 (ORF8) stop codon (Q27stop) mutation). At the same time, the RBD N501Y is thought to increase spike-receptor binding affinity.
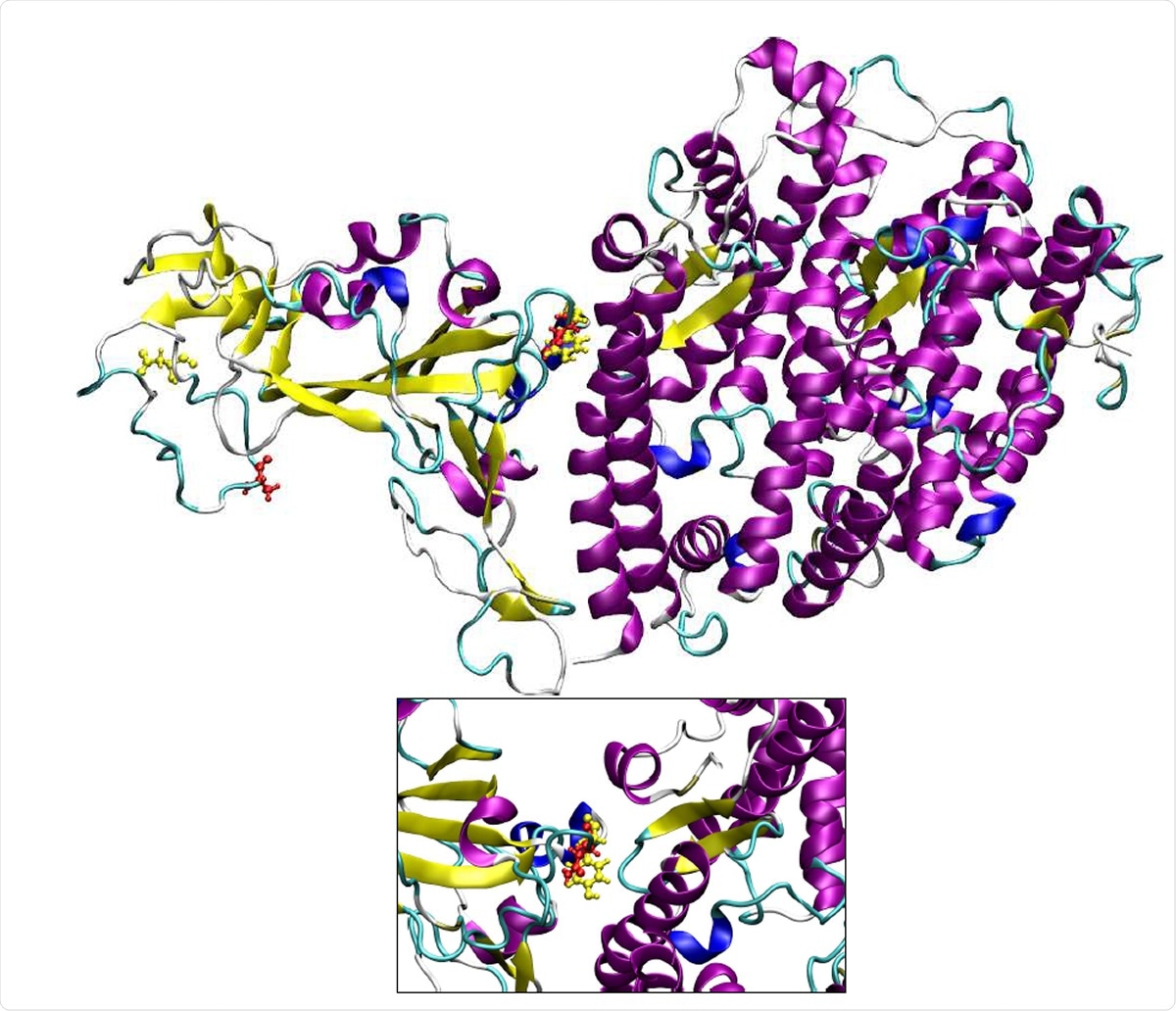
Three dimensional structure the spike protein:hACE-2 complex. The residues that are mutated in B.1.1.7 variant are shown in yellow color.
High number of mutations
The unusually high mutation rate occurring within a short period of time indicates the operation of selection pressure operating on the different virus strains in the same patient following treatment with convalescent plasma.
The researchers offer three explanations for such a phenomenon: prolonged infection with the virus in an immunocompromised patient that allowed evasive mutations to occur at high rates, adaptive mutations that initially occurred in the animal hosts and were transmitted to the human host by zoonotic transmission, and selective pressure brought to bear by antibodies, leading to either direct selection or genetic hitchhiking.
Apart from the effects on the spike's binding affinity to its receptor, the angiotensin-converting enzyme 2 (ACE2), mouse studies indicate increased virulence as well. The double deletion mutation at position 69-70 on the spike protein in the UK variant has also been seen in the Danish Y453F mutant mink strain and the N439K mutation in humans.
The ORF8 mutation inactivates the protein and leads to more mutations downstream, as a result of which the illness may be milder. Other studies suggest a minor effect as a result of this mutation.
Potential consequences
Some suggested implications of these mutations in the UK strain include a 70% increase in transmission and an increase in the reproduction number by 0.4. The gold standard test (reverse transcriptase-polymerase chain reaction, RT PCR) may not recognize the virus RNA if the double deletion H69/V70 occurs. The spike antigen could be altered by the RBD mutations, allowing the virus to escape neutralization by antibodies and lower vaccine efficacy.
Therefore, the current study aimed to understand the various mutations in this strain and how it affects infection, replication, and pathogenesis with SARS-CoV-2.
Study details
The researchers examined the bonds between the spike RBD and the ACE2 receptor, the wild-type spike, and the double mutated spike, the latter containing the N501Y and A570D mutations. The analysis was carried out using molecular dynamics (MD) simulations to determine complex stability over a long time scale.
These studies also yielded computational properties such as the RMSD (root mean square displacement), root mean square fluctuation (RMFS), and hydrogen bond analysis, the last pertaining only to the hydrogen bonds formed between the spike and the ACE2 receptor. The data obtained were used to find the binding free energies using the molecular mechanics-Generalized Born surface area approach (MM-GBSA).
Increased spike-ACE2 interactions
The results show that the number and stability of hydrogen bonds differ between the wild-type and the mutant variant, with the latter resulting in a more significant number of bonds that survive for a more extended period, probably leading to increased spike-ACE2 interactions. In fact, the mutated-spike-ACE2 interaction is higher than the wild-type spike-ACE2 interaction, by ~20 kcal/mol.
The N501Y mutation is found to lower the binding free energies for the ACE-2 receptor by 17-19 kcal/mol. This allows increased interaction between the viral spike and the host cell ACE2, which accounts for the higher infectivity.
The spike mutant has higher stability, by -4 to -10 kcal/mol than the wild-type spike. It also contributes to more significant conformational fluctuations, though this is due to increased flexibility over a small set of residues (527-580). This indicates the effects of the mutation on the structure and dynamics of the ACE2 receptor.
The increased stability of the mutant spike-ACE2 complex formed explains the increased virulence of this variant, as it promotes the spread of the virus between infected cells. The flexibility could cause a mutational escape from antibody-mediated neutralization.
Stability due to increased interactions
The stability is explained by the higher interaction between different pairs of residues, especially TYR449-ASP38, TYR453-HIE34, and TYR501-LYS353, with a greater number of hydrogen bonds between the peptides. Thus, the researchers found that the higher spike-ACE2 affinity was due to hydrogen bonds between pairs of residues not involved directly in the N501Y mutation.
What are the implications?
The mutated spike of the UK variant interacts more strongly and stably with the ACE2 receptor, with lower binding free energies by -20 to -21 kcal/mol. These findings show why this strain is more virulent, with a few pairs of residues interacting more often with a higher number of intermolecular bonds between them.
The decreased free energy of the spike mutant relative to the wild-type spike protein seems to suggest that its suitability for long-term retention and propagation in newly emerging variants. Thus, these computational methods are capable of allowing a better understanding of how mutations affect the interactions within the spike-ACE2 complex.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources