COVID-19 disease is caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 was declared a pandemic by the World Health Organization in March 2020. As of today, there have been over 106 million reported infections of SARS-CoV-2 and more than 2.31 million deaths attributed to COVID-19 disease globally.
The ongoing COVID-19 pandemic emphasizes the importance of molecular surveillance to understand the evolution of the virus and to monitor and plan the epidemiological responses
An epidemiological approach to surveillance aims at finding the source of an outbreak and the nature of transmission chains. Some tools ideal for this purpose are phylogenetic trees, maps, and timelines. However, a pathogen-related approach that focuses on vaccinations, treatments, and resistances needs method-specific visualizations and offers several specific applications. Public health research groups or researchers from universities often study the same pathogens but may still need slightly different visualizations, for example, to analyze different genes, which can necessitate time-consuming adaptions. Hence, easy visualization, rapid analysis, and handy filtering of the latest sequences are vital for this approach.
Making sequence information more easily accessible for epidemiologists and virologists
In a recent bioRxiv* preprint research paper, scientists from Germany discuss how they made sequence information more easily accessible for epidemiologists and virologists. The researchers working together with German public health authorities developed a molecular surveillance tool called CovRadar for the SARS-CoV-2 spike protein. The tool has an analysis pipeline and a web application.
According to the researchers, CovRadar is very flexible and can handle large amounts of data, and users can customize it based on their needs. Both the pipeline and the app can be installed and run on local systems so that access-restricted sequences can be included, and all functionalities are offered at covradar.net.
The coronavirus spike protein has a receptor-binding domain (RBD), which is a target for several vaccine candidates. CovRadar makes it possible to analyze and visualize more than 250,000 sequences.
It first extracts the regions of interest using local alignment and then develops a multiple sequence alignment, infers variants, consensus sequences, and phylogenetic trees and then shows the results in an interactive app, making reporting handy and flexible.
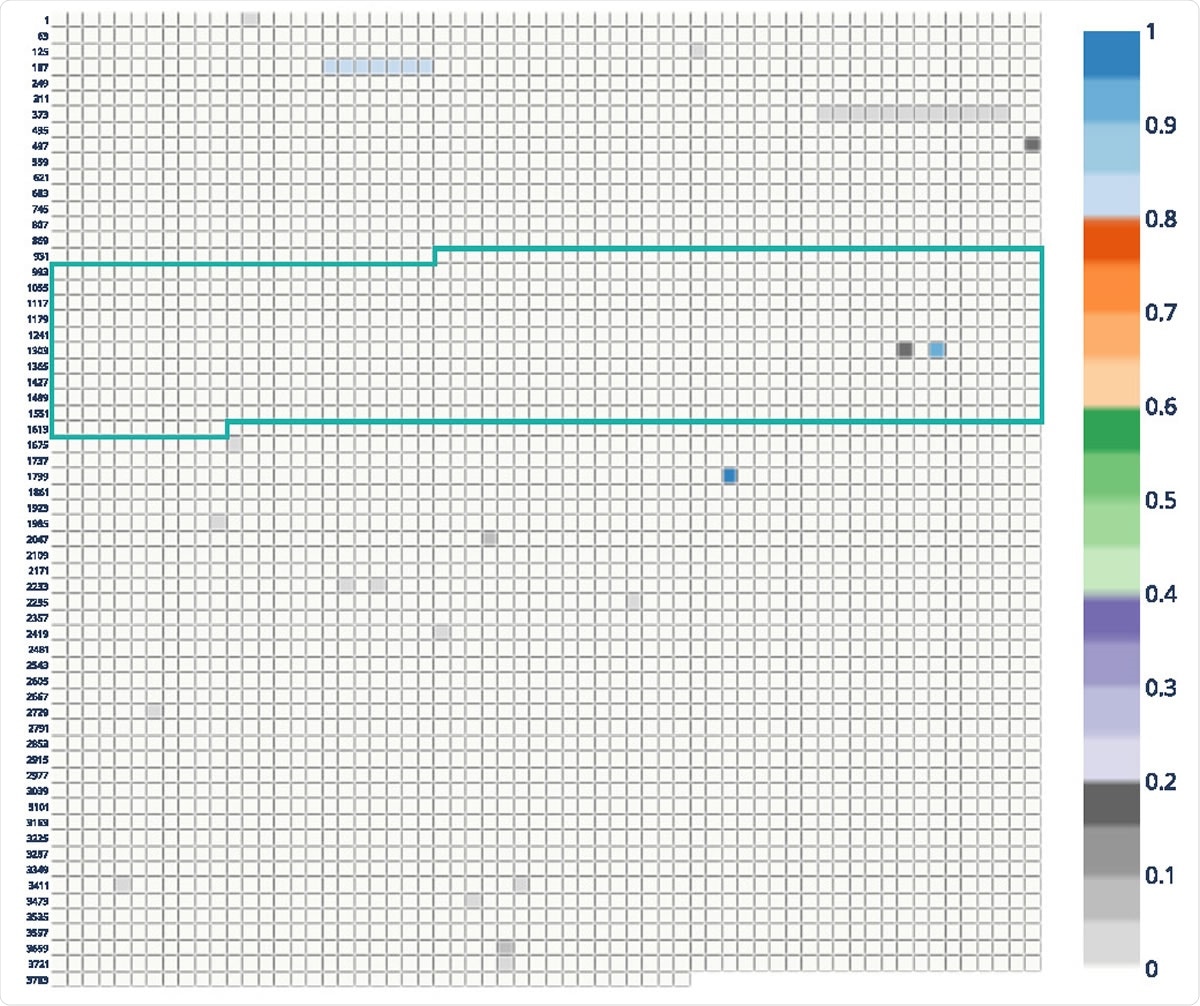
Allele frequency plot of the Danish mink sequences with framed receptor-binding domain (RBD). Each block represents a nucleotide in the MSA of spike gene sequences. Coordinates on the left are related to the MSA position. The plot shows frequencies > 0:8 for nucleotide positions corresponding to codons 68–70, 453 and 614.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
CovRadar has a PDF-like reporting layout with interactive functionalities
The interactive report on the SARS-CoV-2 spike protein is an application written in Flask and Dash and is linked to a MySQL database. The web app layout is similar to a PDF report with interactive functionalities, including filtering, printing, and navigation in the side menu bars. Users can save the tables and plots in the form of excel sheets and PNG, respectively, to use them in other ways. All elements, including plots and tables, are directly editable.
CovRadar can help with situations where zoonotic transmissions with host-specific mutations are possible. Several mutations are currently under discussion to determine if they are related to mink and their significance for the vaccines in development. With the help of CovRadar, the researchers analyzed the positions in the current Danish mink data with high allele frequencies and compared the data with human data both before and after the mink outbreak. They found an increase of alternative alleles mainly at codon positions 453 and 68–70, which could be related to Y435F and del69-70.
CovRadar is accessible free of cost at https://covradar.net. It is open-source code is available at https://gitlab.com/dacs-hpi/covradar.
“CovRadar was designed to assist the molecular surveillance of the Corona spike protein used as target for most vaccine candidates.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
CovRadar: Continuously tracking and filtering SARS-CoV-2 mutations for molecular surveillance, Alice Wittig, Fábio Miranda, Ming Tang, Martin Hölzer, Bernhard Y. Renard, Stephan Fuchs, bioRxiv, 2021.02.03.429146; doi: https://doi.org/10.1101/2021.02.03.429146, https://www.biorxiv.org/content/10.1101/2021.02.03.429146v1
- Peer reviewed and published scientific report.
Johnson, Bryan A., Xuping Xie, Adam L. Bailey, Birte Kalveram, Kumari G. Lokugamage, Antonio Muruato, Jing Zou, et al. 2021. “Loss of Furin Cleavage Site Attenuates SARS-CoV-2 Pathogenesis.” Nature, January, 1–10. https://doi.org/10.1038/s41586-021-03237-4. https://www.nature.com/articles/s41586-021-03237-4.