A team of scientists from the United States has recently explored the prevalence and transmission dynamics of the B.1.1.7 variant (UK variant) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the United States. The study reveals that because of 35-45% increased transmissibility, B.1.1.7 may soon become the US's dominant variant. The study is currently available on the medRxiv* preprint server.
As of February 9, 2021, there have been over 106 million confirmed infections of SARS-CoV-2, including 2.33 million deaths, reported to the World Health Organization (WHO).
The concern over the pandemic is increasing globally particularly because of the emergence of new variants of SARS-CoV-2, which are more infectious and may be more lethal than the wildtype virus.
During the later part of the pandemic in late 2020, the B.1.1.7 variant containing spike N501Y mutation has emerged in the UK and rapidly became the predominant variant in the UK and in other European countries.
Similar to the previously identified D614G variant, B.1.1.7 variant's transmissibility is 40 – 70% higher than other SARS-CoV-2 variants identified globally. Moreover, a growing pool of preliminary studies has suggested that the B.1.1.7 variant may be more lethal than other viral variants. Infection with this variant may increase the mortality rate by 30%.
In the current study, the scientists have investigated the prevalence and transmissibility of the B.1.1.7 variant in the US.
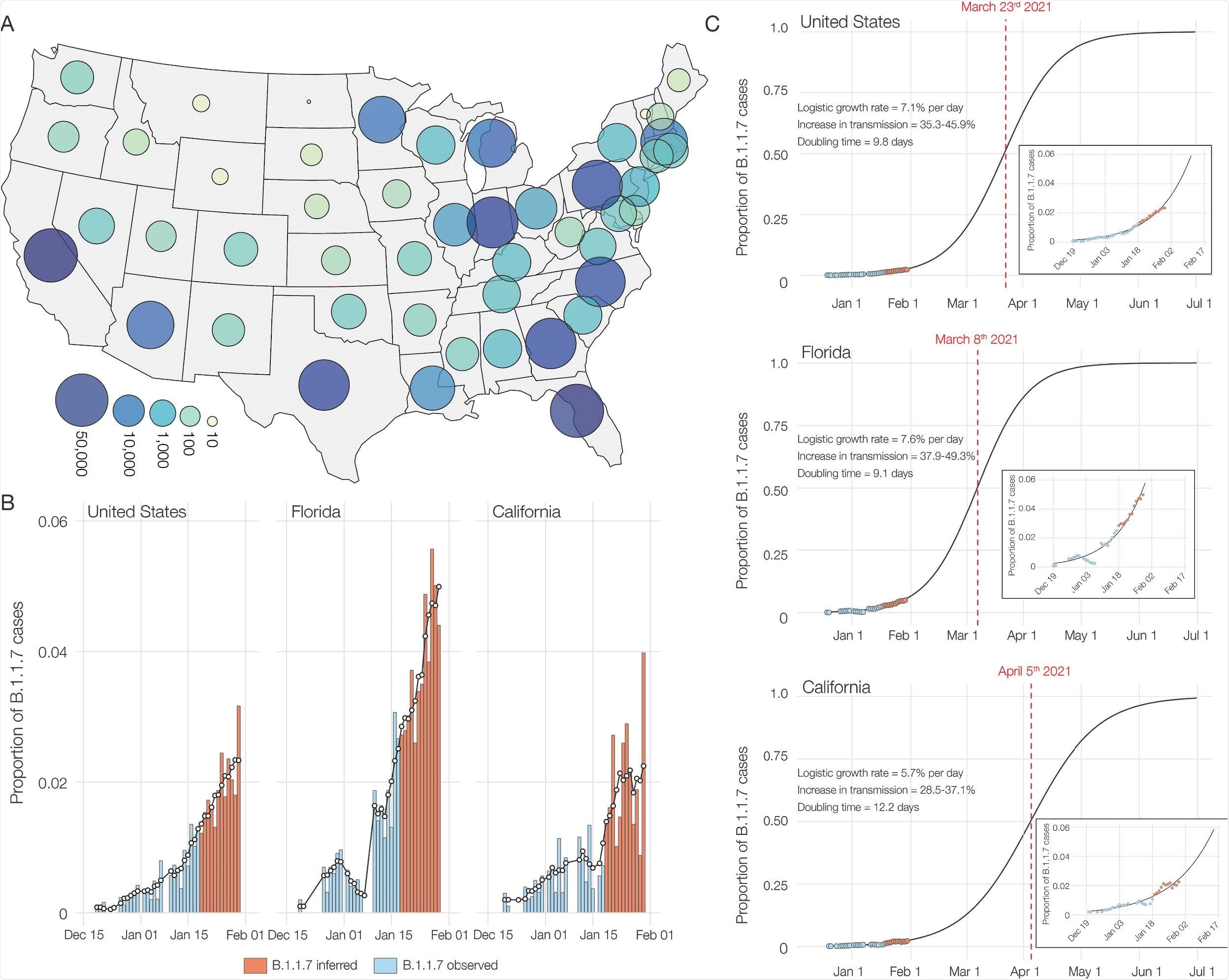
SGTF and B.1.1.7 in SARS-CoV-2 tests at Helix since December 15, 2020. (A) Map of contiguous states in the USA with each bubble representing the number of positive tests from each state. (B) Estimated proportion of B.1.1.7 in total number of positive tests with Cq(N gene) < 27, in the U.S., California and Florida from December 15th, 2020 to January 30th, 2021. The proportion of B.1.1.7 samples was estimated using: (Observed B.1.1.7 sequences/Sequenced SGTF samples) * (Positive tests with SGTF/Total positive tests). Due to the lag in sequencing, the average proportion of B.1.1.7 sequences in sequenced samples with SGTF from the last five days (January 13-18) was used to infer the proportion of B.1.1.7 cases in total positive tests for the January 19-30 time period between. The black line shows the 5-day rolling average of the estimated proportion of B.1.1.7 in total positives. (C) Logistic growth curves fit to the rolling average of the estimated proportion of B.1.1.7 in total positives for the U.S., Florida and California. The predicted time when the estimated proportion of B.1.1.7 cases crosses 0.5 is indicated in red.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study design
The spike gene-target failure (SGTF) in quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)-based diagnosis of SARS-CoV-2 infection was utilized to determine the fraction of B.1.1.7 variant in tested specimens.
When mutations are present at the target probe regions, qRT-PCR method can be used to predict the presence of viral variants with genetic differences. In tested specimens, the presence of 69-70 deletion, which is a signature mutation in the B.1.1.7 variant, can be characterized by SGTF.
A total of 460 SGTF specimens collected between December 2020 and January 2021 were sequenced to estimate the prevalence of the B.1.1.7 variant in the US.
Important observations
While examining the prevalence of SGTF in SARS-CoV-2-positive specimens, the scientists observed that in October 2020, the fraction of SGTF was as low as 0.2%, which increased gradually afterward. An increase in SGTF fraction from 0.8% to 4.2% occurred between the first week and last week of January 2021.
By the end of January 2021, the B.1.1.7 variant's prevalence was 3.6% in the US. Regarding nationwide distribution, they observed a significant variation in SGTF fractions across the nation.
To firmly investigate the B.1.1.7 prevalence, they sequenced all SGTF samples and observed that of 460 SGTF specimens, the B.1.1.7 variant was present in 209 specimens collected from ten US states. Moreover, they sequenced three additional B.1.1.7 genomes from California. Of these 212 B.1.1.7-positive specimens, 96 were obtained from California and 87 from Florida.
Regarding the population-level prevalence of B.1.1.7 variant, they observed that by the end of January 2021, about 2.1% of all confirmed COVID-19 cases in the US were caused by the B.1.1.7 variant.
Using a logistic growth model, they estimated 35 – 45% increased transmissibility of the B.1.1.7 variant in the US, with a doubling time of about 9.8 days.
By conducting time-aware Bayesian phylodynamic analyses, they observed that multiple independent entry events were responsible for the emergence of the B.1.1.7 variant in the US, starting as early as late November 2020.
By estimating the time to the most recent common ancestor, they noticed that the local transmission of B.1.1.7 variant between different US states might have started since December 2020, permitting the rapid transmission of the B.1.1.7 variant at least 30 US states by January 2021.
Study significance
The study reveals that currently, in the US, the B.1.1.7 variant's prevalence is relatively low. However, given the significantly higher transmissibility, B.1.1.7 may soon become the US's dominant variant.
Moreover, the study indicates that multiple international entry events are responsible for the emergence of B.1.1.7 variant in the US, followed by rapid spreading across the nation via community transmission. The scientists suggest that concerned authorities should immediately take vital preventive measures to minimize COVID-19 morbidity and mortality.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources