Proteases are enzymes that catalyze the break-down of proteins by hydrolysis and are key to the infection process of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The spike protein that initially interacts with the ACE2 receptor on the surface of host cells must undergo cleavage to initiate a conformational change that allows entry into the cell. Proteolysis can be facilitated, and has been identified to take place, at several stages: by membrane-bound proteases belonging to the host cell, such as TMPRSS2; extracellular proteases, such as furin; or within an endosome following endocytosis by cathepsins B and L, allowing the virus to escape into the cytoplasm.
.jpg)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Though the above-mentioned proteases have been confirmed to be involved in SARS-CoV-2 spike protein activation, others may still remain unidentified. A research paper recently uploaded to the preprint server bioRxiv* by Platt et al. (17th Feb 2021) aimed to identify these enzymes by computational screening and molecular docking simulations, with cathepsins B, K, L, S, and V being highlighted as probable leads. Interestingly, several preexisting conditions associated with poor outcomes in COVID-19 patients are also associated with elevated levels of these cathepsins, including diabetes and cardiovascular disease. Additional experimental analyses by the generation of a recombinant spike protein and incubation with the enzymes of interest demonstrated the computational model's validity.
What methods were used to identify the enzymes?
The group began by analyzing the SARS-CoV-2 spike protein using a program named Protease-Ase Cleavages from MEROPS ANalyzed Specificities (PACMANS), which identifies probable cleavage site locations by searching for particular amino acid sequences. The program also scores and ranks the cleavage sites based on affinity with the chosen enzyme, allowing the group to perform additional docking simulations on these specific sites.
As mentioned above, Furin and cathepsins B and L have already been implicated in spike protein cleavage, and this process confirmed their involvement. Furin had the highest-rated cleavage site amongst the tested enzymes, shown to be at the S1/S2 subunit site that causes the necessary conformational change for membrane fusion. Other enzymes of interest with cleavage sites in the S1/S2 region included cathepsins B and K, with the others mainly targeting regions of the S1 or S2 domains specifically, or the heptad repeat domains of the spike protein.
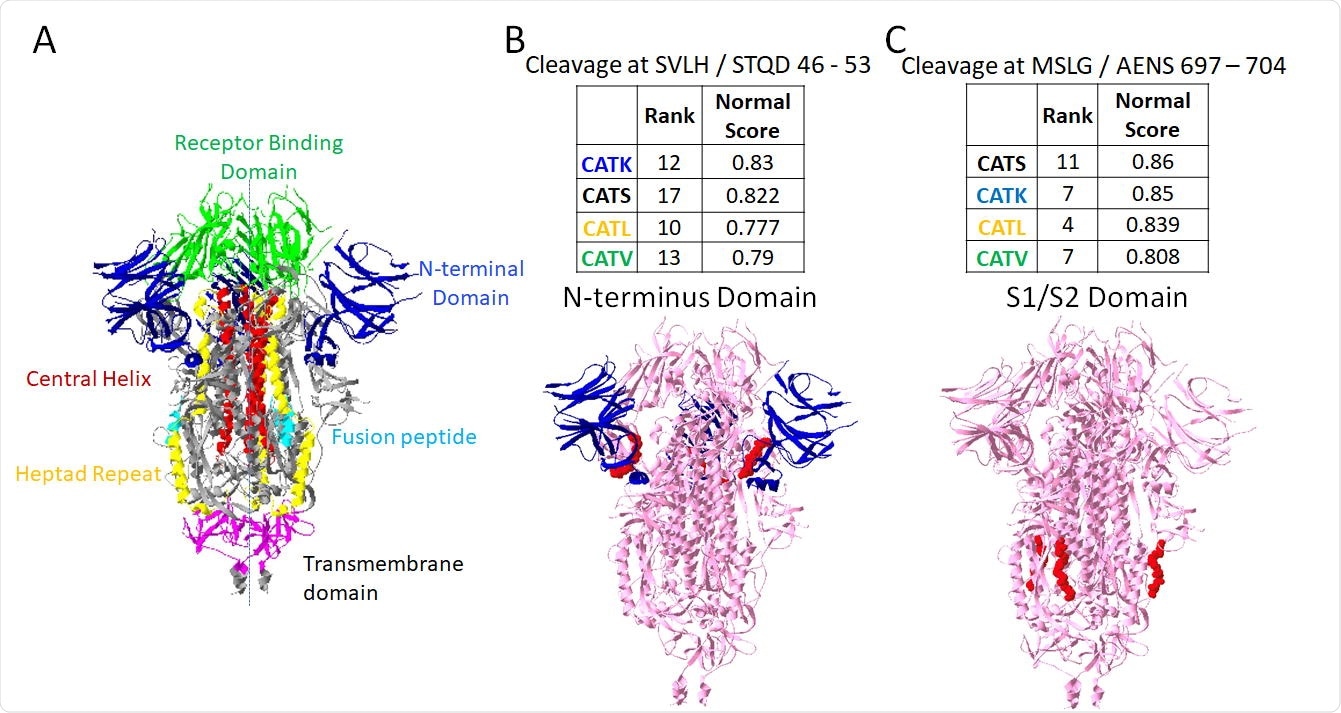
Putative sites susceptible to cleavage by four cathepsins. From PACMANS analysis, there were two amino acid sequences susceptible to cleavage by four cathepsins as indicated by high normalized scores and relatively high-rank orders. From the three-dimensional model of spike protein, the domains are color-coded, (A) and susceptible sequences are highlighted in red. (B) Cleavage after H49 is in the Nterminus domain (blue) of spike protein, and (C) cleavage after G700 was even more highly ranked and cleaves in the S1/S2 domain.
PACMANS only analyzes the sequence of amino acids without considering the three-dimensional structure of the protein. Thus, the enzymes of interest (furin and cathepsins B, K, L, S, and V) were simulated in a molecular docking program against the spike protein in both open and closed conformations. The closeness of the active site of the enzyme with the cleavage site was assessed in three dimensions, with cathepsins L and K coming closest at around 0.3 nm, with the furthest still being only 1.1 nm, generally close enough to initiate binding.
A recombinant form of the full-length spike protein was produced and incubated with the enzymes for varied periods of time, and the cleavage fragments were visualized. Each cathepsin produced unique cleavage fragments that corroborate with the computational findings. Interestingly, the enzymes already confirmed to be involved in the cleavage of the spike protein (cathepsins B and L) actually generated the fewest unique immunodetectable products, with cathepsins K and V generating the most.
What is the implication?
The group hypothesizes that beyond the activating cleavage events observed around the S1/S2 site, multiple simultaneous cleavage events in the N-terminal and receptor-binding domains could have an inactivating effect, being destabilizing protein conformation and preventing cell entry. The fragments released from the N-terminal domain following cleavage here were identified, and it was confirmed that their removal could cause destabilization and unfolding of the spike protein.
This study highlights the varied role these proteases play in the activation, deactivation, cell entry, and virulence of the SARS-CoV-2 virus and presents a methodology by which other enzymes could be screened for greater insight into the progression of COVID-19 disease.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Multiple Sites on SARS-CoV-2 Spike Protein are Susceptible to Proteolysis by Cathepsins B, K, L, S, and V Keval Bollavaram, Tiffanie H. Leeman, Maggie W. Lee, Akhil Kulkarni, Sophia G. Upshaw, Jiabei Yang, Hannah Song, Manu O. Platt, bioRxiv, 2021.02.17.431617; doi: https://doi.org/10.1101/2021.02.17.431617, https://www.biorxiv.org/content/10.1101/2021.02.17.431617v1
- Peer reviewed and published scientific report.
Bollavaram, Keval, Tiffanie H. Leeman, Maggie W. Lee, Akhil Kulkarni, Sophia G. Upshaw, Jiabei Yang, Hannah Song, and Manu O. Platt. 2021. “Multiple Sites on SARS‐CoV ‐2 Spike Protein Are Susceptible to Proteolysis by Cathepsins B, K, L, S, and V.” Protein Science 30 (6): 1131–43. https://doi.org/10.1002/pro.4073. https://onlinelibrary.wiley.com/doi/10.1002/pro.4073.