New data from researchers led by Majid Kazemian from Purdue University refutes claims from recent studies suggesting the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could create a host-virus chimeric RNA. Their team found SARS-CoV-2 does not combine with the human genome.
The study “Host-virus chimeric events in SARS-CoV-2 infected cells are infrequent and artifactual” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Low probability of HVC events
Given the high percentage of error associated with creating an RNA sequencing library, the researchers hypothesized that HVC events were improbable.
The researchers reanalyzed three RNA-sequencing datasets from 57 patients with SARS-CoV-2 and 64 samples of in vitro SARS-CoV-2 infected cells.
Results showed SARS-CoV-2 infected about 20%-70% of Calu-3 and A549-ACE2 cells.
They next looked for any combined viral and human genome instances, which would indicate an HVC event. While the Calu-3 and A549-ACE2 cells showed the highest percentages of chimeric read, this was from 0.5-1% of all read sequences.
When looking at whether some viral genome regions were more inclined to fuse, the team found chimeric reads occurred more on the 3’ end of the SARS-CoV-2 genome, where the viral N protein is located.
Similar results were also seen when looking at SARS-CoV-2 RNA sequencing, suggesting a fusion of the viral and human genome is unlikely to happen.
The last experiment involved amplifying SARS-CoV-2 sequences from the RNA of infected host cells. While the enriched viral sequences should have increased the number of chimeric events, they did not.
This suggests a chimeric event is limited by the number of RNA fragments with the 3’ end is available.
The researchers write:
“This is consistent with a stochastic model in which chimeric events are dependent on the availability of template RNA, i.e., the more viral RNA fragments present, the higher the chance of participation in chimeric events.”
A similar observation was made in virus-infected cells collected from patients with COVID-19 infection. While a low chance of chimeric events had occurred, a correlation was observed between the number of viral genes expressed and the number of chimeric events.
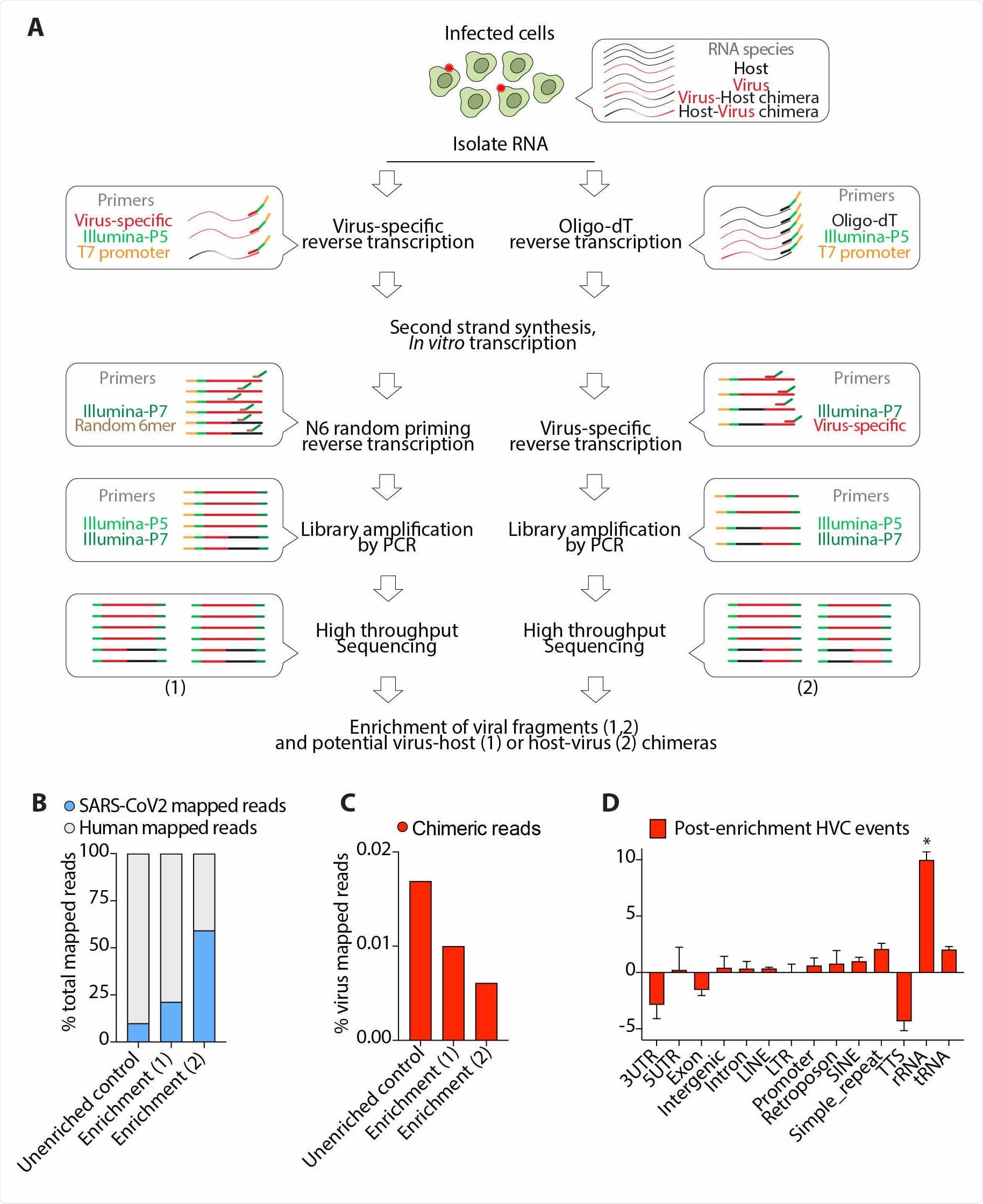
Experimental enrichment for viral-containing fragments does not enrich for HVC events. (A) Schematic presentation of viral RNA enrichment from infected host cells. Cellular RNA from infected cells comprises host RNA, viral RNA and presumably any fusion RNA between virus and host. A pool of oligo probes that are specific to SARS-CoV2 was used in a series of reverse transcription, in vitro transcription and PCR amplification steps to amplify viral RNAs and potential virus-host (1) or host-virus (2) chimeras (see Methods). (B) Viral reads in the indicated libraries from SARS-CoV2-infected Calu-3 cells as a proportion of the total reads mapped to the chimeric genome. (C) HVC reads in the indicated libraries from SARS-CoV2- infected Calu-3 cells as a proportion of the total reads mapped to the SARS-CoV2 genome. (D) Distribution of genomic features in the human segment of HVC events detected after enrichment for viral-containing transcripts. * p<0.05 by Wilcoxon test.
Viral-human hybrids are not reproducible
The researchers next decided to rule out HVCs as a common feature in the life cycle of the SARS-CoV-2 virus. The team read RNA-sequencing data from a series of independent studies to find evidence of either exon-exon splicing junctions in HVCs events that could be transmitted to another.
Results showed no HVC event that was reproducible across data sets.
To confirm their findings, they looked at the proportion of unique reads in the RNA-sequencing dataset that covered an HVC junction. They reasoned that the greater number of reads, the more likely an HVC was an isolated event.
Of all the HVC events that were read, only 2-15% had reads spanning their junction. “This is in clear contrast to 90-95% and 40-70% of known and novel splicing events, respectively, that have more than one supporting read.”
Based on the low chance of reproducibility, the authors conclude HVCs are likely nothing more than non-biological artifacts.
Artifacts likely an error during RNA transcription
How the artifacts came to be is still a mystery, but the researchers postulate they may have been created due to the reverse transcriptase enzyme, which is prone to error.
To test this, they compared the genomes of fruit flies with spiked-in RNA. About 5% of reads were attributed to the fruit fly genome. They then mapped the RNA that could be considered a hybrid between human and fruit fly RNA.
“Since there is no actual possibility of biological fusion events between host and spiked-in RNAs, we considered any chimeric reads identified as artifactual.”
Indeed, approximately 1% of the mapped area could be mistaken as chimeric events. This is similar to the less than 1% of artifacts observed in SARS-CoV-2 infected cells.
Amplifying viral RNA does not increase HVC events
There was a possibility that the researchers could not have been detected chimeric events because the number of viral reads was low. To circumvent this issue, the team developed a technique to enhance the frequency of viral RNA with the idea that it should also enhance the number of HVC events.
However, the number of HVC events did not change with their chance of occurrence at less than 0.05%.
Based on the given evidence, the authors strongly suggest any indication of HVC events from infected SARS-CoV-2 cells are likely artifacts/noise from RNA sequencing.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Yan B, et al. Host-virus chimeric events in SARS-CoV2 infected cells are infrequent and artifactual. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.17.431704, https://www.biorxiv.org/content/10.1101/2021.02.17.431704v1
- Peer reviewed and published scientific report.
Yan, Bingyu, Srishti Chakravorty, Carmen Mirabelli, Luopin Wang, Jorge L. Trujillo-Ochoa, Daniel Chauss, Dhaneshwar Kumar, et al. 2021. “Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual.” Edited by Colin R. Parrish. Journal of Virology 95 (15). https://doi.org/10.1128/jvi.00294-21. https://journals.asm.org/doi/10.1128/JVI.00294-21.