Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interacts with angiotensin-converting enzyme 2 (ACE2) receptors on the surface of host cells via the receptor-binding domain (RBD) of the viruses spike protein.
The spike protein structure and its domains have been cataloged since early in the pandemic, and this information has proven vital in the development of diagnostic procedures and vaccines. However, the specific structural characteristics and the surface glycan profile are highly dependent on synthesis conditions. Those that are biotechnologically produced are likely to differ from their natural form.
A research paper recently uploaded to the preprint server bioRxiv* by Dominguez-Vega et al. (23rd Feb, 2021) describes an in-depth structural analysis of the RBD of the spike protein expressed in mammalian Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK293) cells, providing a new understanding of protein structure and function.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Why is this important?
Recombinant spike proteins have previously been produced in full and short forms to develop neutralizing antibodies and vaccines by several routes, including by human embryonic kidney cells. When produced, however, these proteins were noted to bear differing glycan structures, which could potentially alter the way the spike protein and RBD interact with ACE2 and antibodies. Several researchers have noted that recombinantly produced spike proteins may differ based on their source, the length of protein expressed, and any post-translational modifications that may take place. Careful replication of the true spike protein is essential for effective vaccine and neutralizing antibody creation.
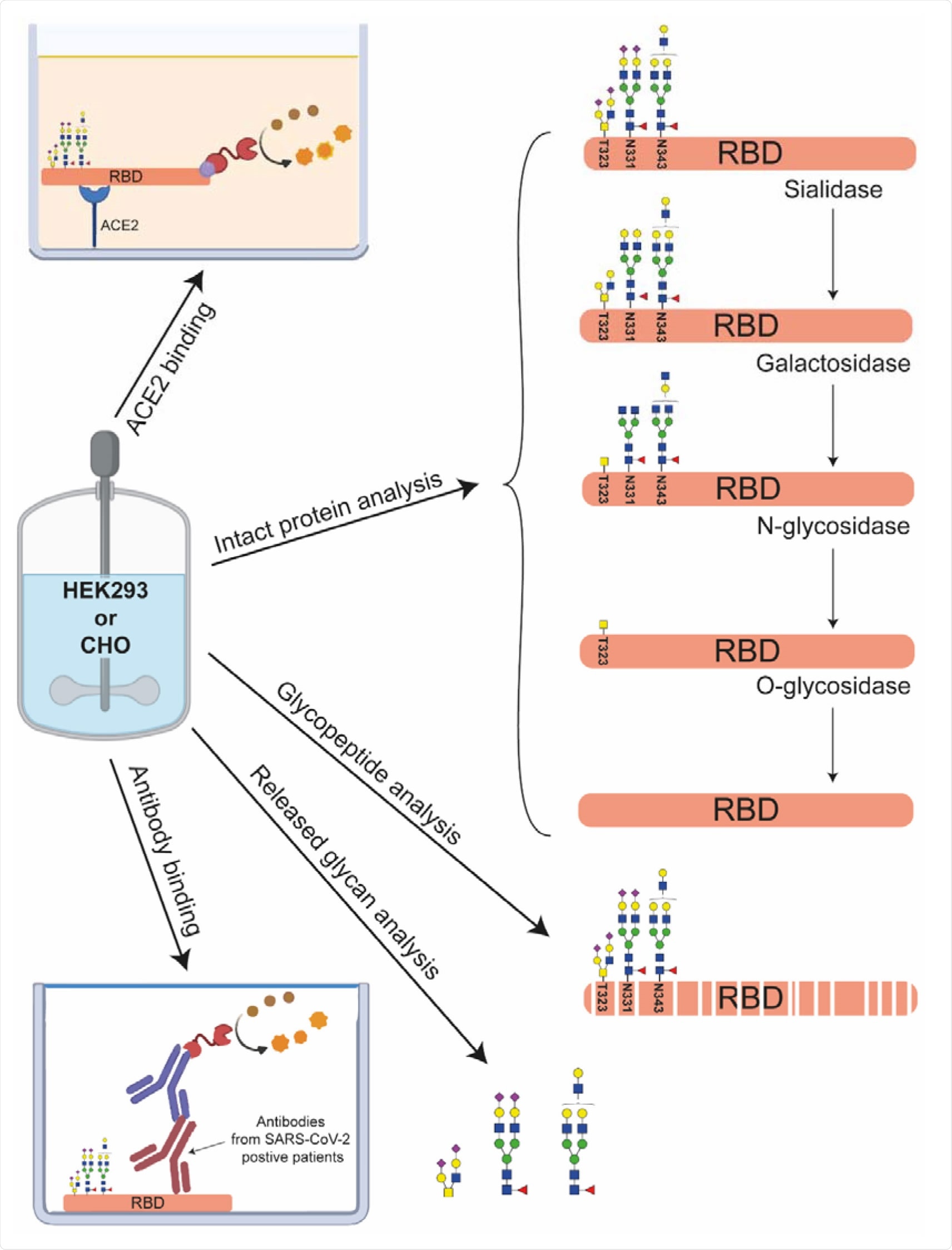
Multilevel characterization of RBDs produced in HEK293 or CHO cells. Next to intact protein analysis using CE-MS and MALDI-ISD-MS the samples are structurally characterized by glycan dissection, glycopeptide analysis and released glycan analysis. Their functional characterization was performed by measuring their binding characteristics to ACE2 and anti-SARS-CoV-2 antibodies.
How did the RBDs differ?
The group performed an in-depth structural analysis of two commercially available recombinant RBDs produced by both CHO and HEK293 cells, beginning with capillary electrophoresis-mass spectrometry (CE-MS) after deglycosylation of the RBD to establish protein sequences and any difference in mass between the RBDs without regarding glycans. Those produced by both CHO and HEK293 cells were on average 119 Da larger than the protein's theoretical mass, explained by the presence of a free cysteine amino acid that becomes cysteinylated during antibody production.
Matrix-assisted laser desorption ionization in-source-decay (MALDI-ISD) mass spectrometry, which is often used to fragment proteins into ions directly in the ion source, was performed, and it was found that HEK293 cells had more differences from the expected in the number and type of N-terminal proteins. Pyroglutamic acid forms from glutamic acid residues and 52% of the sequence was found to be affected this way. HEK293 cells have a slightly different sequence that results in additional glutamine at the N-terminus after cleavage, explaining the mass shift of 111 Da compared with CHO RBD after taking into account pyroglutamic acid formation.
N-glycans and O-glycans were individually released and analyzed by CE-MS and MALDI-ISD MS. HEK293 had larger heterogeneity, and more glycans were sialylated, with many of those being di- or tri- sialylated, which affects the isoelectric point of the protein.
Antibody binding assays were performed with the glycosylated and non-glycosylated RBDs against both anti-SARS-CoV-2 IgG antibodies collected from patient sera and the ACE2 receptor. Both RBDs performed similarly in each case. The non-glycosylated RBDs exhibiting a halved affinity towards the ACE2 receptor, as expected from earlier reports and theorized to be due to the conformational stability provided to the RBD by glycans. The affinity of either RBD towards ACE2 was greater than that of either the full spike protein or RBD-containing subunit, most likely due to lessened steric hindrance around the binding site and thus greater access to the receptor.
Overall, CHO cells were less sialylated but bore more high-antennary and LacNAc repeat structures, while HEK293 cells had higher sialylation and antenna fucosylation. Additionally, the group discovered a new O-glycosylation site that is not present in the full-form spike protein and could have implications in receptor binding.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Structural and functional characterization of SARS-CoV-2 RBD domains produced in mammalian cells Christoph Gstoettner, Tao Zhang, Anja Resemann, Sophia Ruben, Stuart Pengelley, Detlev Suckau, Tim Welsink, Manfred Wuhrer, Elena Dominguez-Vega bioRxiv 2021.02.23.432424; doi: https://doi.org/10.1101/2021.02.23.432424, https://www.biorxiv.org/content/10.1101/2021.02.23.432424v1
- Peer reviewed and published scientific report.
Gstöttner, Christoph, Tao Zhang, Anja Resemann, Sophia Ruben, Stuart Pengelley, Detlev Suckau, Tim Welsink, Manfred Wuhrer, and Elena Domínguez-Vega. 2021. “Structural and Functional Characterization of SARS-CoV-2 RBD Domains Produced in Mammalian Cells.” Analytical Chemistry 93 (17): 6839–47. https://doi.org/10.1021/acs.analchem.1c00893. https://pubs.acs.org/doi/10.1021/acs.analchem.1c00893.