Researchers in Switzerland and Germany have identified host cell factors required for coronavirus replication that could serve as targets for treatment with clinically-approved drugs.
The team found that several autophagy-related genes were common host defense factors required for the replication of both endemic and emerging coronaviruses.
These coronaviruses include the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic.
Autophagy – cellular response to stressors such as hypoxia or infection – involves the recycling of proteins and organelles to maintain homeostasis. Various trafficking pathways enable the transportation of cytoplasmic material to the lysosome, where it is destroyed.
Among the autophagy-related genes were three immunophilins – high affinity-receptor proteins that specifically bind to certain immunosuppressive agents.
Furthermore, inhibition of the immunophilins with the clinically-approved drugs Cyclosporin A and Alisporivir resulted in dose-dependent reduction of coronavirus replication in primary human nasal epithelial cells.
The study was conducted by a team from the Institute of Virology and Immunology in Bern and Mittelhäusern, Switzerland and Ruhr-Universität Bochum in Germany
“Overall, we identified host factors that are crucial for coronavirus replication and demonstrate that these factors constitute potential targets for therapeutic intervention by clinically approved drugs,” writes Volker Thiel and the team.
A pre-print version of the paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Three highly pathogenic coronaviruses have emerged over the last two decades
The last two decades have seen the emergence of three highly pathogenic coronaviruses, including the SARS-CoV virus responsible for the 2002-2004 SARS outbreaks, the Middle Eastern respiratory syndrome coronavirus (MERS-CoV) that emerged in 2012 and, most recently, the SARS-CoV-2 virus that causes COVID-19.
The severe risk these outbreaks have posed to human health over a relatively short period has highlighted the importance of developing effective approaches to treating both current coronavirus infections and those that could emerge in the future.
Coronaviruses rely on host dependency factors
Coronaviruses rely on cellular host factors – termed host dependency factors (HDFs) – for viral entry, replication and survival.
“The identification of HDFs is therefore important for understanding essential host-virus interactions required for successful viral replication and providing a framework to guide the development of new pharmacological strategies for the treatment of coronavirus infections,” says Thiel and colleagues.
One hallmark process that occurs during coronavirus replication is extensive virus-induced remodeling of host endomembranes to form double-membrane vesicles (DMVs) that are targeted by viral replication and transcription complexes.
“However, the host factors that are required for the formation of these structures remain elusive,” says the team.
What did the researchers do?
The researchers conducted two independent genome-wide loss-of-function CRISPR screens to identify HDFs required for the replication of both endemic and emerging coronaviruses.
The knockout screens were performed in Huh7 cells infected with the highly pathogenic MERS-CoV and with human coronavirus 229E (HCoV-229E) – a less pathogenic endemic coronavirus that generally only causes mild respiratory symptoms.
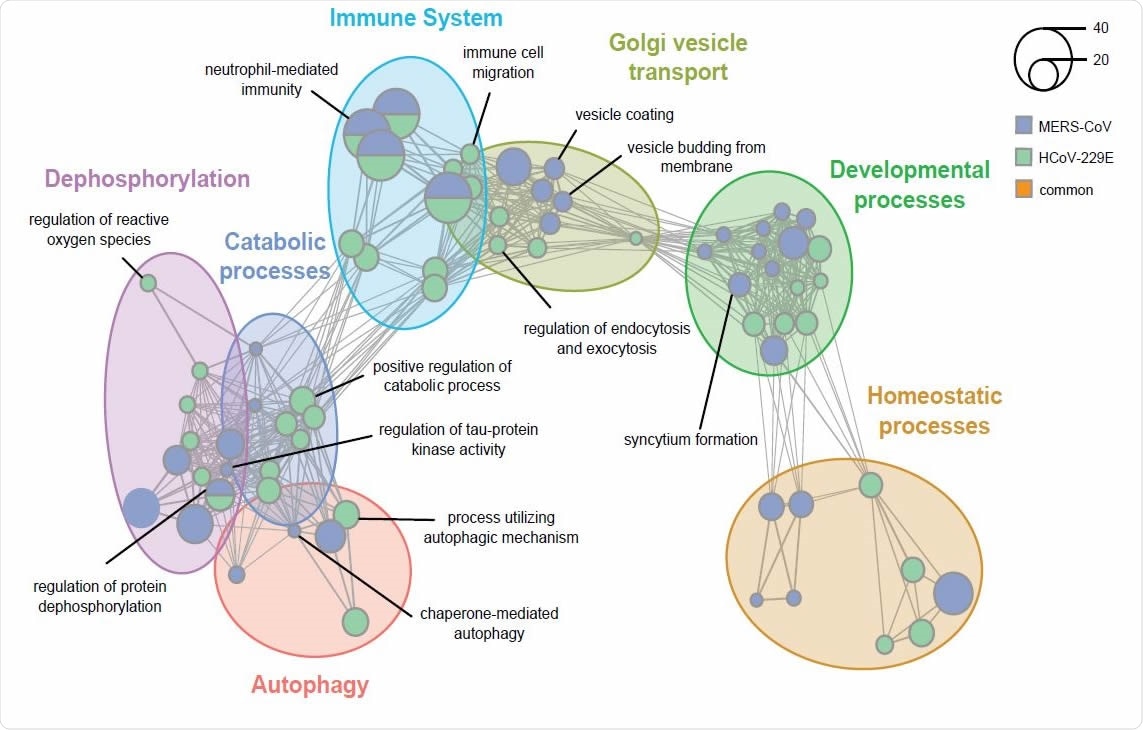
Enrichment analysis uncovers host biological networks crucial for CoV replication. (A) Enrichment map summarizing major host biological networks co-opted by CoVs during infection. Gene Ontology (GO) enrichment analysis was performed using hits from both MERS-CoV and HCoV-229E CRISPR screens and filtered to contain conserved representative GO terms and genes. Each node represents an individual GO term and nodes that are functionally related cluster together into a larger network. Node size reflects number of significantly enriched genes in the node and color indicates the CoV screen for which the node was significant.
What did the study find?
The team identified multiple virus-specific and conserved HDFs, including several that are required for replication of SARS-CoV-2.
The study revealed that several autophagy-related genes, including the immunophilins FK506 binding protein 8 (FKBP8), transmembrane protein 41B (TMEM41B), and membrane integral NOTCH2-associated receptor 1 (MINAR1) were common HDFs.
The researchers say that the interaction between autophagy components and coronaviruses in the context of replication has been considered for some time because parts of the autophagy process share similarities with the process of DMV formation.
However, “studies investigating the possible involvement of the early autophagy machinery in the conversion of host membranes into DMVs reached conflicting conclusions,” says Thiel and colleagues.
“Another possibility is that single components of the autophagic machinery may be hijacked by coronaviruses independently of their activity in autophagic processing,” they add.
The team says that irrespective of the precise underlying mechanism, the results suggest that FKBP8, TMEM41B, and MINAR1 represent potential therapeutic targets.
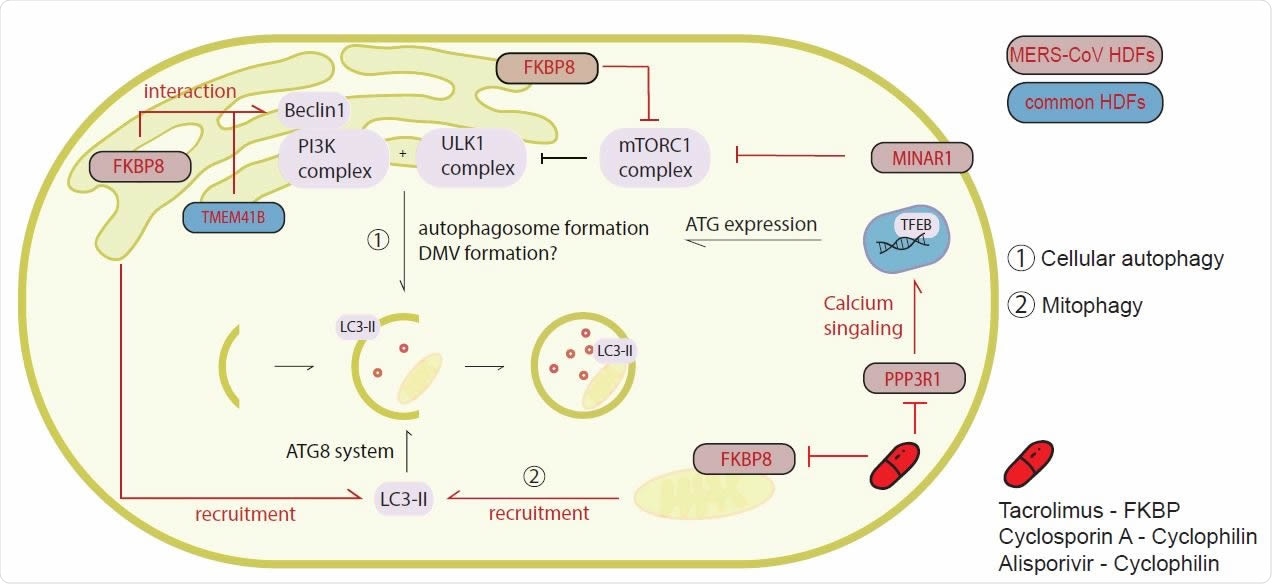
CoV HDFs are interactors of the autophagy pathway but do not depend on autophagy for replication. (A) Upon starvation, the mTORC1 complex is blocked and activation of the PI3K complex, as well as the ULK1 complex leads to the initiation of phagophore formation, as an initial step in the autophagy pathway. MERS-CoV and HCoV503 229E top scoring CRISPR knockout screen hits FKBP8, MINAR1, TMEM41B and VMP1 are involved in this early pathway. Furthermore, the ATG8 system containing among others LC3, which is recruited by VPM1 or FBKP8 is necessary for targeting cellular cargo to the autophagosome. PPP3R1 is upregulated and initiates TFEB translocalization to the nucleus, where it catalyzes transcription of ATGs. MERS-CoV or conserved host dependency factors (HDFs) are indicated in respective colors. Inhibitor intervention in this pathway is shown in red.
Targeting the immunophilins with clinically-approved drugs
Next, the researchers showed that inhibition of the immunophilin family with the clinically-approved and well-tolerated drugs Tacrolimus, Cyclosporin A and Alisporivir reduced the replication of MERS-CoV, SARS-CoV, and SARS-CoV-2 in a dose-dependent manner.
However, the team noted that while Huh7 cells are valuable for studying coronaviruses, they are likely less effective at capturing important aspects of infection than primary human airway epithelial cells.
To address this limitation, the researchers also tested the drugs in primary human nasal epithelial cell cultures.
This revealed that Cyclosporin A and Alisporivir potently inhibited SARS-CoV-2 replication at concentrations known to be achievable and efficacious in patients.
“Overall, the genes and pathways identified in our coronavirus screens expand the current repertoire of essential HDFs required for replication that can be exploited to identify novel therapeutic targets for host-directed therapies against both existing and future emerging CoVs,” writes Thiel and colleagues.
“Together these findings depict a promising path towards the repurposing of Cyclosporin A and Alisporivir as COVID-19 treatment options,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Thiel V, et al. A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.02.24.432634, https://www.biorxiv.org/content/10.1101/2021.02.24.432634v1
- Peer reviewed and published scientific report.
Kratzel, Annika, Jenna N. Kelly, Philip V’kovski, Jasmine Portmann, Yannick Brüggemann, Daniel Todt, Nadine Ebert, et al. 2021. “A Genome-Wide CRISPR Screen Identifies Interactors of the Autophagy Pathway as Conserved Coronavirus Targets.” Edited by Ken Cadwell. PLOS Biology 19 (12): e3001490. https://doi.org/10.1371/journal.pbio.3001490. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001490.