Researchers at the New York State Department of Health in the United States have said concerning mutations within the B.1.526 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has recently become prevalent in New York State warrant further monitoring.
The SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to sweep the globe posing a threat to public health and the economy worldwide.
Erica Lasek-Nesselquist and colleagues say that as well as acquiring the E484K mutation, which is known to help the virus escape neutralizing antibodies, the B.1.526 lineage already harbors a D235G substitution that could also help to reduce the efficacy of neutralizing antibodies.
Both the E484K and D235G mutations are located in a viral protein called spike – the main surface structure that SARS-CoV-2 uses to bind to and enter host cells.
As the initial stage of the infection process, the receptor-binding domain (RBD) of the spike attaches to the host cell receptor angiotensin-converting enzyme 2 (ACE2). This spike RBD–ACE2 interaction is the main target of neutralizing antibodies following infection with SARS-CoV-2.
Furthermore, another mutation – S477N – has arisen within the B.1.526 lineage that is associated with increased ACE2 receptor binding affinity and resistance to neutralization by monoclonal antibodies.
The researchers say the combination of E484K, S477N and D253G mutations that might confer immune escape warrants further monitoring.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.
Several variants of concern have emerged over recent months
Several SARS-CoV-2 variants of concern have emerged internationally over recent months. These include the B.1.1.7 variant identified in the UK in September 2020, the B.1.351 variant identified in South Africa in October 2020 and the P.1 variant detected in Brazil in December 2020.
All three lineages harbor a set of mutations in the viral spike protein that is crucial to the initial stage of infection.
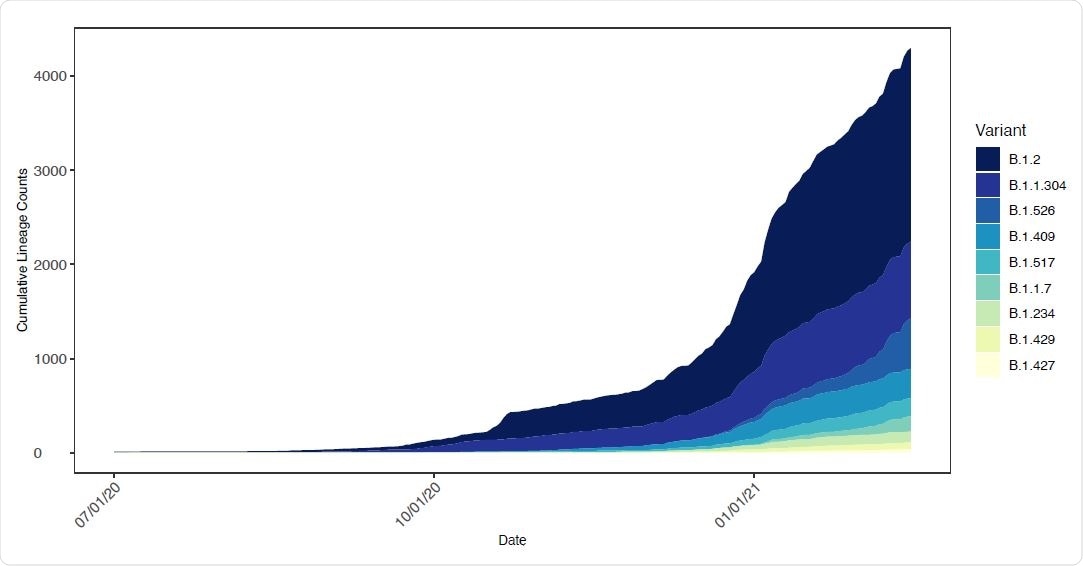
Relative frequency of SARS-CoV-2 lineages with E484K mutations in New York State. The number of SARS CoV-2 genomes assigned to lineages containing E484K as a function of time. The lineages included contained at least one genome with an E484K mutation in the New York State dataset.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The mutations K417N and E484K found in B.1351 and P.1 are of particular concern since they significantly reduce the neutralization activity of monoclonal antibodies and convalescent plasma.
However, “while surveillance efforts have increased in the US, the country has not yet met the recommended sequencing of 5% of all COVID-19 cases in order to detect variants before they reach high prevalence in the population,” says Pata and the team.
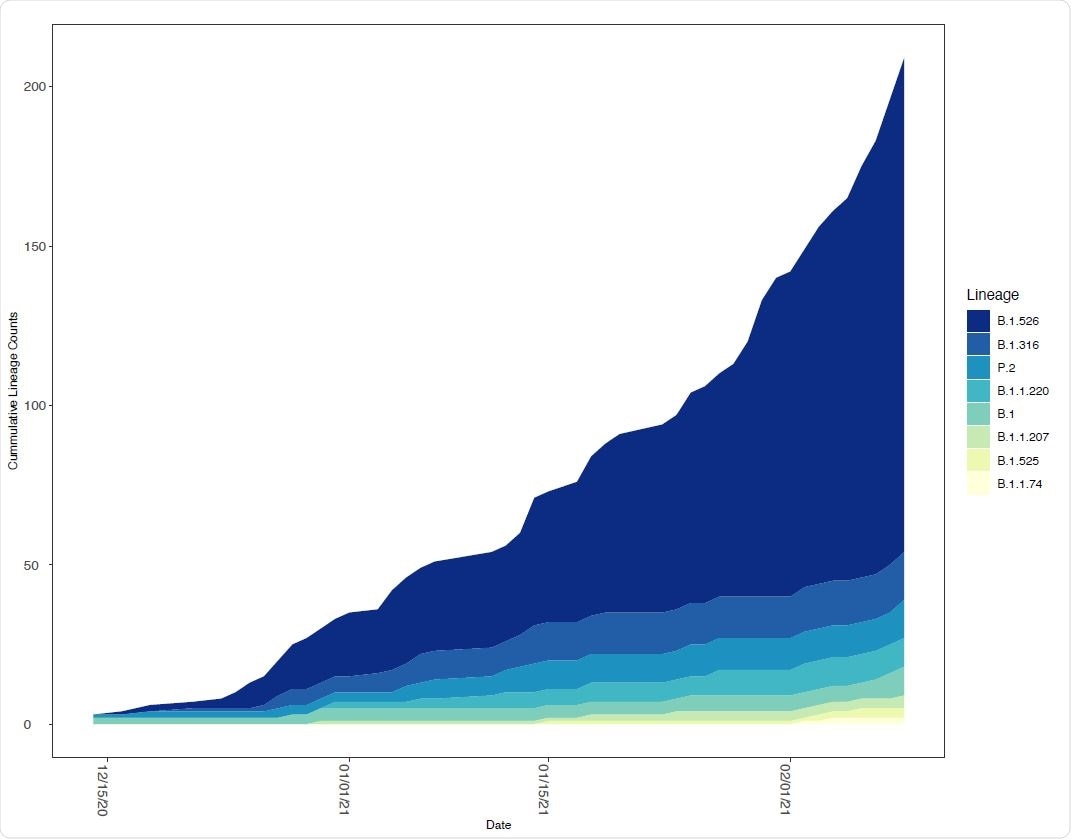
Changing frequency of B.1.526 and other lineages in New York State. The number of genomes assigned to select SARS-CoV-2 lineages circulating in New York State as a function of time. Only lineages with 10 or more genomes from samples collected within the past 14 days were included.
What did the researchers do?
Pata and colleagues analyzed mutations present in more than 10,000 SARS-CoV-2 genomes from New York that were cataloged by NextClade.
The team reports the emergence of the E484K mutation within the B.1.526 lineage that has become 26 times more prevalent across New York State in just over a month.
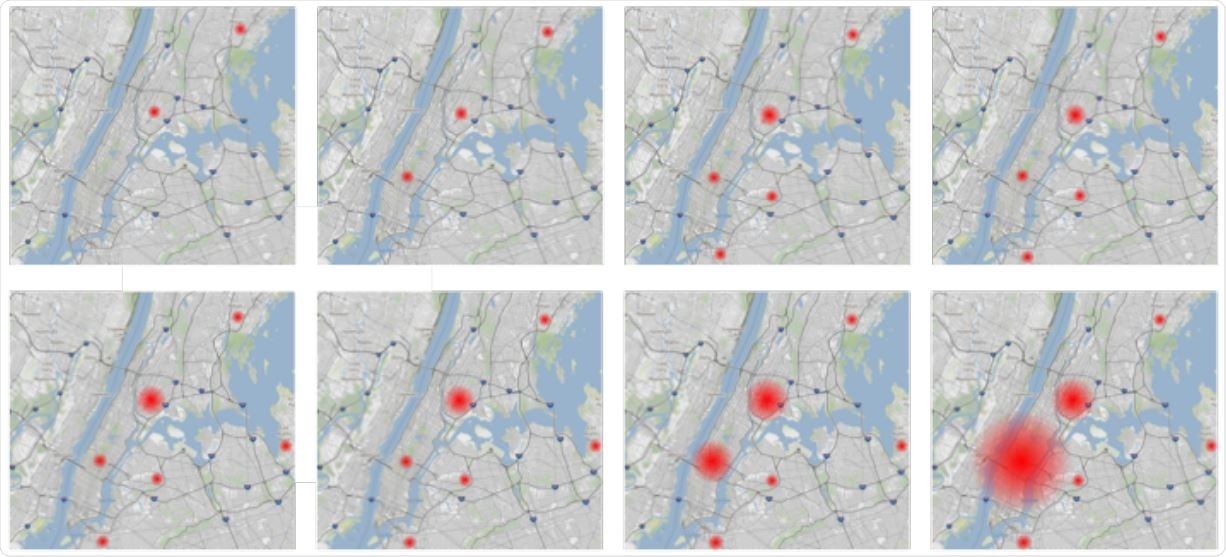
Increase in Frequency of the B.1.526 E484K variant in the NYC metropolitan area. Starting from the top left, each panel encompasses a seven-day period from 2020-12-16 to 2021-02-03 showing the increase of B.1.526 484K variants (red dots). The red dots are proportional to the total number of variants found in an area.
The researchers say that since the E484 mutation is located in the spike RBD – at the interface with the ACE2 receptor – it could increase binding affinity and contribute to the already dramatically increased rates of B.1.526 484K variants in New York.
“Other mutations present in B.1.526 likely do not explain the increase in transmission,” writes the team.
What about the D235G and S477N mutations?
The B.1.526 lineage also harbors five other amino acid substitutions in the spike protein. One of these substitutions – D253G – occurs in the “supersite” loop of the N-terminal domain (NTD) of spike and is a key residue that interacts with most NTD-targeting neutralizing antibodies. This D235G mutation could therefore help to reduce the efficacy of neutralizing antibodies.
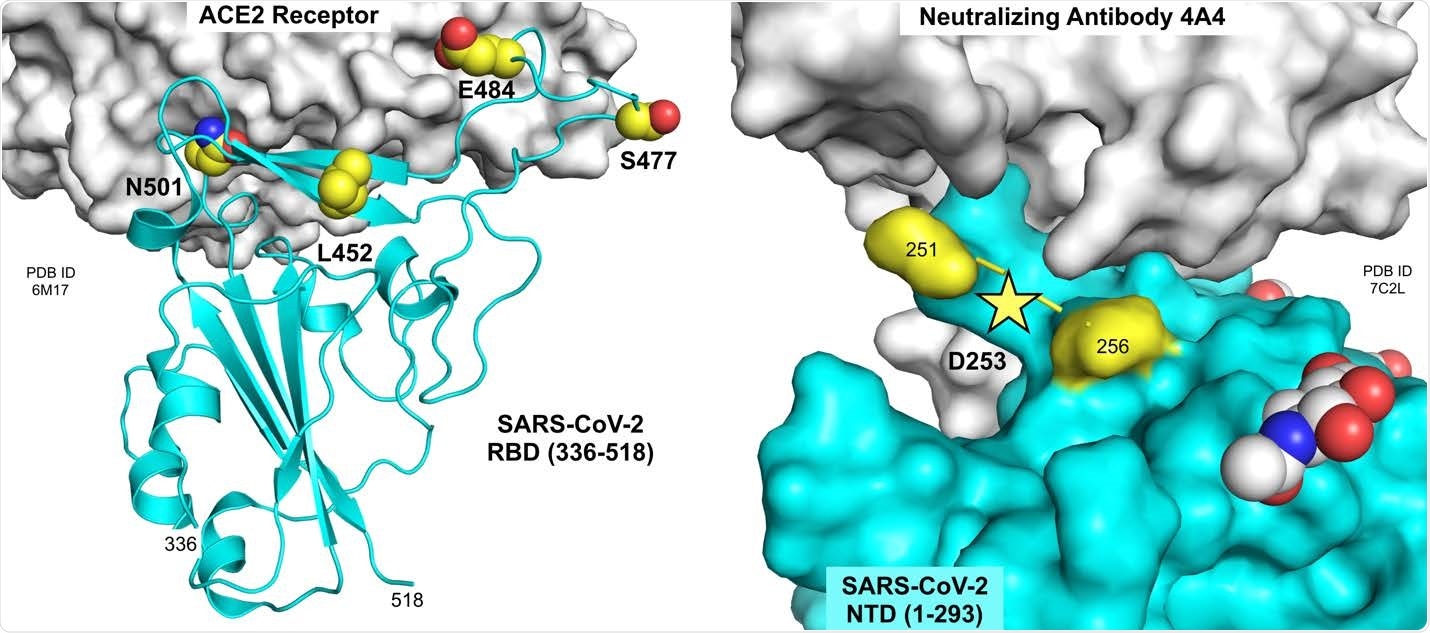
Residues mutated in B.1.526 spike protein. Residues 452, 477, 484 and 501 are located at or near the interface of the RBD with the ACE2 receptor (left panel; PDB ID 6M17, Yan et al., 2020). Residue 253 is located in a small disordered loop in the NTD near the interface with neutralizing antibody 4A4 (right panel; PDB ID 7C2L, Chi et al., 2020). Figure prepared using the PyMOL Molecular Graphics System v2.3.5 (Schrödinger, LLC).
Furthermore, another mutation – S477N – that is associated with increased ACE2 receptor binding affinity and resistance to antibody neutralization has emerged within the B.1.526 lineage.
Now, the E484K and S477N spike variants currently comprise the majority of B.1.526 cases, says Pata and colleagues.
“We note the emergence of new variants in the B.1.526 lineage, which now harbor E484K or S477N mutations in addition to the already present D253G change,” they write.
The mutations have helped make B.1.526 the fifth most abundant lineage
The researchers say that, collectively, these variants now represent 81% of B.1.526 SARS-CoV-2 genomes in New York State, helping to make it the fifth most abundant lineage since its detection in mid-December.
“The combination of E484K or S477N with a D253G mutation that might confer immune escape and the increased number of COVID-19 cases associated with these variants warrants further monitoring,” concludes the team.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.