Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel coronavirus, emerged into the human population in late 2019, in Wuhan, China. During the ensuing worldwide COVID-19 pandemic, already more than 2.6 million lives have been lost due to the virus. The spread of SARS-CoV-2 has thus far been extremely challenging to contain. One of the reasons for the setback is the prevalence of pre-symptomatic and asymptomatic infected individuals who can, unknowingly, transmit the virus to others. Thereby, these individuals can be regarded as “super spreaders” of the SARS-CoV-2 virus and are also super-spreaders of COVID-19 disease.
In a new study published on the medRxiv* preprint server, researchers analyzed viral loads from pre-symptomatic and asymptomatic individuals in an extensive university surveillance program. Saliva samples were collected from the University of Colorado Boulder (USA) campus for the period of August to December 2020. A total of 72,500 samples were obtained. Out of these, 1,405 positive COVID-19 cases were detected. Viral loads were evaluated in saliva because it is considered the most reliable biospecimen to identify carriers of the virus.
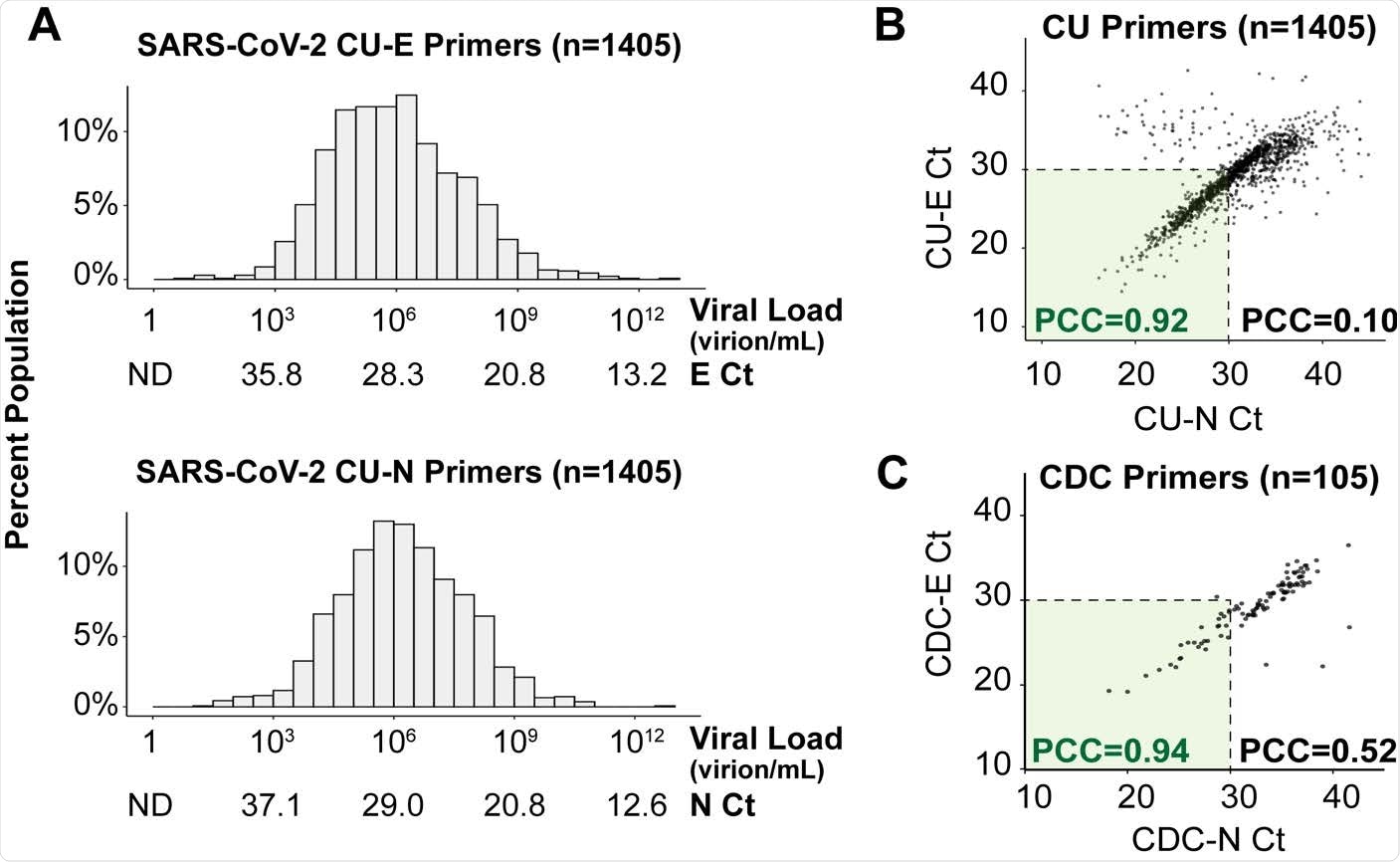
Saliva viral load distribution within our campus population. A) Viral load distribution within the 1,405 positive samples identified on campus during the Fall semester of 2020. Each histogram shows Ct values obtained using TaqMan primer/probe sets targeting either the E gene (“CU-E”) or the N gene (“CU-N”) of SARS-CoV-2. The horizontal axes are labeled with both the cycle threshold values (Ct) and the corresponding viral loads calculated from the standard curve for each primer set. ND = no data, as the viral load is below the RT-qPCR detection limit. B) The Ct values resulting from the two primer sets in panel A are highly correlated, especially in samples with high viral loads (Ct value lower than 30). Pearson correlation coefficients (PCC) are shown within and beyond the Ct=30 arbitrary cutoff. C) For 105 of the SARS-CoV-2 positive saliva samples, we ran RT-qPCR side-by-side with 8 different primer sets commonly used in SARS-CoV-2 diagnostic tests. Here, we show the same analysis as in panel B, except with the U.S. Centers for Disease Control’s (CDC) primers targeting the E and N genes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Saliva is also the most likely medium for the transmission of the respiratory SARS-CoV-2 pathogen. A unique feature of this dataset is that all SARS-CoV-2-positive persons reported no symptoms during the time of saliva collection. Hence, these individuals were infected with SARS-CoV-2 during the time of the sample collection but were either asymptomatic or pre-symptomatic. The data set revealed a wide variance in viral loads between individuals. The research confirmed a tiny percentage of infected persons carry the high-threshold level of infectious virions to be able to transmit the virus.
The saliva samples collected for this study were tested using quantitative RT-PCR. One of the most important highlights of this study is that most circulating virions prevail in a small number of individuals of the infected population. Their findings corroborate those obtained by other researchers who have conducted similar studies. More work has to be conducted to study the correlation between the transmission mode and the viral loads.
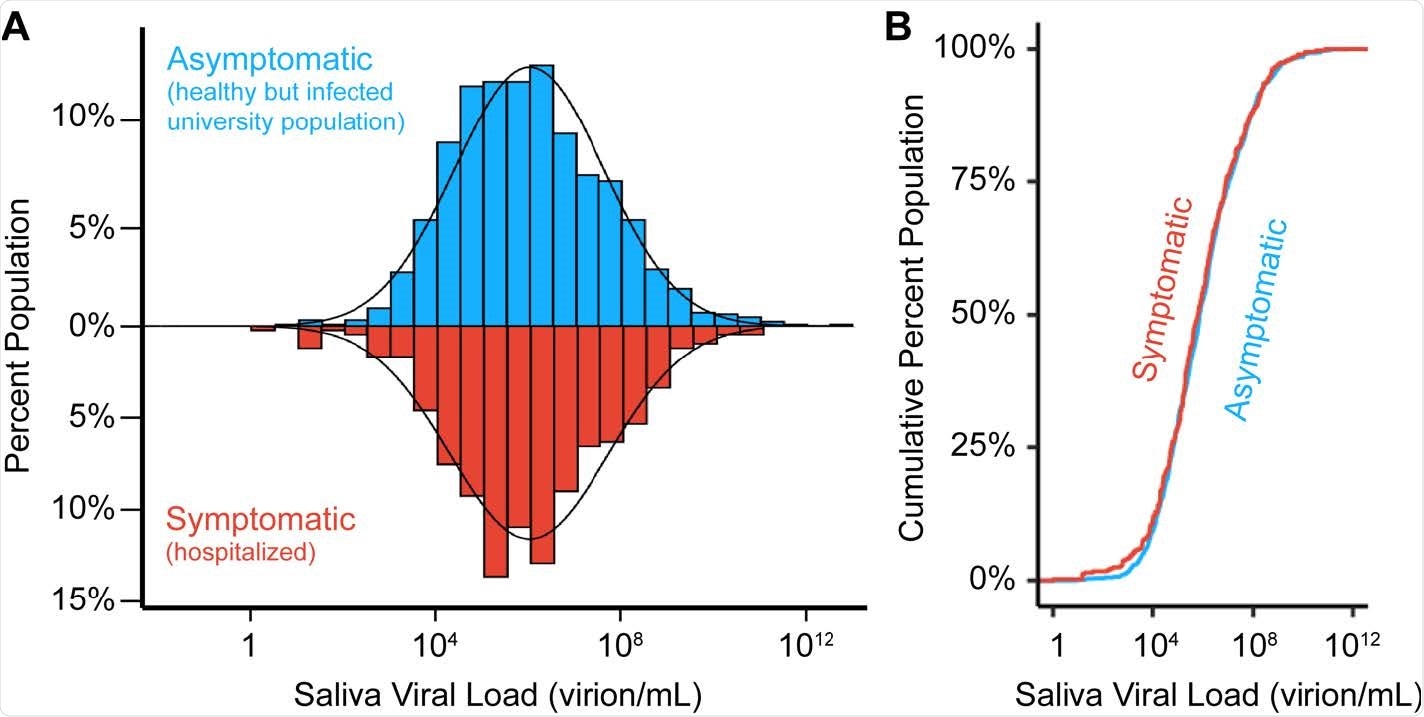
Viral load distributions are similar in asymptomatic and symptomatic populations. A) A histogram of saliva viral loads in our asymptomatic campus population (N=1405, blue) compared to the same histogram of saliva viral loads from symptomatic (N=404, red) individuals. A log-normal probability density function is fitted onto the two distributions given the population mean and standard deviation. B) Empirical cumulative distribution functions (ECDFs) of saliva viral load in pre-symptomatic (N=1405, blue) and symptomatic (N=404, red) populations. The similarity of the two ECDFs were assessed with the Kolmogorov-Smirnov test, which resulted in D statistic = 0.04, and p-value = 0.72.
The current study implies that individuals with high levels of viral loads may be “super-spreaders.” Previous studies conducted in countries such as China and Spain have shown that higher viral loads in individuals increase the probability of transmitting the virus to others. A similar result was obtained in the current study, where the probability of transmission increased between the roommates of the University of Colorado campus students.
The current study conducts contact tracing analyses and supports the claim that viral super-carriers spread infection at a higher rate. The data revealed that around 80-90% of infections are spread by 10-20% of infected individuals. Additionally, this study also suggests that only a small fraction of individuals infected with SARS-CoV-2 contain a viral load level that facilitates active transmission. Similar episodes were reported between roommates, schoolmates, and household members. The researchers found that just 2% of individuals harbored 90% of the circulating virions. Further, 99% of community-circulating virions are accounted for by just 12% of the asymptomatic and 14% of the symptomatic population.
![A small percentage of individuals are viral super-carriers. The histograms shown (right Y axes) are the same as were shown in figure 2. Starting from the left of each histogram (i.e. those individuals with the highest viral loads), we calculated the accumulative percentage of total virions as a function of saliva viral load based on the probability density function of the distribution (blue and red lines, and left Y axes). In both asymptomatic (blue line) and symptomatic populations (red line), the portion of population that harbors 90% and 99% of the circulating virus is highlighted by the dashed lines. We estimate that only about 50% of individuals who test positive for the virus actually harbor live virus, based on the observation that live virus has rarely been isolated from samples with viral loads <106 virions per mL [28,30–35]. For context, typical limits of detection of three major SARS-CoV-2 testing paradigms are shown. RT-qPCR on purified RNA is the most sensitive, with RT-LAMP and antigen tests performed directly on biological fluids being less sensitive. However, all testing paradigms will capture virtually all infectious individuals and virions, in pre-symptomatic and symptomatic populations alike. A small percentage of individuals are viral super-carriers. The histograms shown (right Y axes) are the same as were shown in figure 2. Starting from the left of each histogram (i.e. those individuals with the highest viral loads), we calculated the accumulative percentage of total virions as a function of saliva viral load based on the probability density function of the distribution (blue and red lines, and left Y axes). In both asymptomatic (blue line) and symptomatic populations (red line), the portion of population that harbors 90% and 99% of the circulating virus is highlighted by the dashed lines. We estimate that only about 50% of individuals who test positive for the virus actually harbor live virus, based on the observation that live virus has rarely been isolated from samples with viral loads <106 virions per mL [28,30–35]. For context, typical limits of detection of three major SARS-CoV-2 testing paradigms are shown. RT-qPCR on purified RNA is the most sensitive, with RT-LAMP and antigen tests performed directly on biological fluids being less sensitive. However, all testing paradigms will capture virtually all infectious individuals and virions, in pre-symptomatic and symptomatic populations alike.](https://www.news-medical.net/image-handler/picture/2021/3/ds21252250v1.jpg)
A small percentage of individuals are viral super-carriers. The histograms shown (right Y axes) are the same as were shown in figure 2. Starting from the left of each histogram (i.e. those individuals with the highest viral loads), we calculated the accumulative percentage of total virions as a function of saliva viral load based on the probability density function of the distribution (blue and red lines, and left Y axes). In both asymptomatic (blue line) and symptomatic populations (red line), the portion of population that harbors 90% and 99% of the circulating virus is highlighted by the dashed lines. We estimate that only about 50% of individuals who test positive for the virus actually harbor live virus, based on the observation that live virus has rarely been isolated from samples with viral loads <106 virions per mL [28,30–35]. For context, typical limits of detection of three major SARS-CoV-2 testing paradigms are shown. RT-qPCR on purified RNA is the most sensitive, with RT-LAMP and antigen tests performed directly on biological fluids being less sensitive. However, all testing paradigms will capture virtually all infectious individuals and virions, in pre-symptomatic and symptomatic populations alike.
Researchers have perceived that one of the potential reasons for the difference in the viral load between infected individuals is that they are tested at different viral infection phases. However, the longitudinal analyses of the viral load of every infected individual show that peak viral load varies significantly between individuals. Thereby, researchers concluded that the viral load produced by each individual is different. This may be due to the difference in their immune response. Additionally, factors such as variation in the host’s characteristics related to virus replication, e.g., ACE2 and/or the type of virus variant infecting the individual, play a vital role in the variation of viral load in infected individuals.
The current research shows that the concentration of a high level of viral load is only prevalent in a very small fraction of the population at a given time. Thereby, increasing the frequency of the COVID-19 test or community screening for the identification of viral super-carriers within the pre-symptomatic and asymptomatic stages can play a significant role in containing the pandemic. As mentioned above, if these super-carriers are not identified, then the pandemic will prolong as the virus transmission between individuals continues.
Modeling approaches show that one of the significant factors in screening the community for SARS-CoV-2 infection is the speed with which infected people receive their test results. The turnaround time will determine how fast the asymptomatic persons can be isolated from the others. In other words, a delay in the turnaround time will rapidly spread the infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities, Qing Yang, Tassa K. Saldi, Erika Lasda, Carolyn J. Decker, Camille L. Paige, Denise Muhlrad, Patrick K. Gonzales, Morgan R. Fink, Kimngan L. Tat, Cole R. Hager, Jack C. Davis, Christopher D. Ozeroff, Nicholas R. Meyerson, Stephen K. Clark, Will T. Fattor, Alison R. Gilchrist, Arturo Barbachano-Guerrero, Emma R. Worden-Sapper, Sharon S. Wu, Gloria R. Brisson, Matthew B. McQueen, Robin D. Dowell, Leslie Leinwand, Roy Parker, Sara L. Sawyer, medRxiv, 2021.03.01.21252250; doi: https://doi.org/10.1101/2021.03.01.21252250, https://www.medrxiv.org/content/10.1101/2021.03.01.21252250v1
- Peer reviewed and published scientific report.
Yang, Q., Saldi, T. K., Gonzales, P. K., Lasda, E., Decker, C. J., Tat, K. L., Fink, M. R., Hager, C. R., Davis, J. C., Ozeroff, C. D., Muhlrad, D., Clark, S. K., Fattor, W. T., Meyerson, N. R., Paige, C. L., Gilchrist, A. R., Barbachano-Guerrero, A., Worden-Sapper, E. R., Wu, S. S., & Brisson, G. R. (2021). Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities. Proceedings of the National Academy of Sciences of the United States of America, 118(21), e2104547118. https://doi.org/10.1073/pnas.2104547118, https://www.pnas.org/doi/full/10.1073/pnas.2104547118