The coronavirus disease, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), first emerged in December 2019 in Wuhan City, China. Scientists believe that the virus jumped from an intermediate animal host to humans.
To date, the virus has sickened more than 117 million people and killed over 2.61 million worldwide. It is, therefore, crucial to determine how the virus spreads and evolves to develop vaccines and therapies.
Researchers at Colorado State University and the University of Pennsylvania School of Veterinary Medicine found that SARS-CoV-2 variants rapidly emerge in animal hosts, hinting at the potential risk for human reinfection.
The study, published on the preprint server bioRxiv*, highlights the need to study viral evolution and pathogenesis in human and animal hosts. This could help prevent future outbreaks that may mimic the magnitude of the current COVID-19 pandemic.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study background
Over the years, cross-species transmission events or spillovers, which challenge pathogens to survive in new host environments, result in species-species adaptations.
The severe acute respiratory syndrome outbreak in 2002, the Middle East respiratory syndrome in Saudi Arabia in 2012, and the current COVID-19 pandemic, all emerged from a spillover event. Though bats were most likely the reservoir of coronaviruses, the intermediate hosts include civet cats, camels, and pangolins, among others.
Cross-species transmission events are rare, and in natural settings, they typically fail to result in an epidemic spread. In contrast to most species, humans move globally and regularly come into contact with domestic and peridomestic animals. Thus, when a novel virus spreads through human populations, this may cause a spillback into a wide range of companion and wild animals, which has occurred during the current coronavirus pandemic. Given that reverse zoonosis has been reported in animals like cats and dogs from households where people are infected with SARS-CoV-2, the risk of transmitting the virus to animals is plausible.
The study
The study aimed to assess the evolution of SARS-CoV-2 during three rounds of expansion of strain USA-WA1/2020 in Vero E6 cells, followed by changes happening during the primary experimental infection in four mammalian hosts.
The researchers compared variants of cell culture-expanded SARS-CoV-2 inoculum and virus recovered from four species following exposure to the virus. They also compared variant proportions, insertions, and deletions occurring in genomes of SARS-CoV-2 obtained from dogs, cats, hamsters, and ferrets.
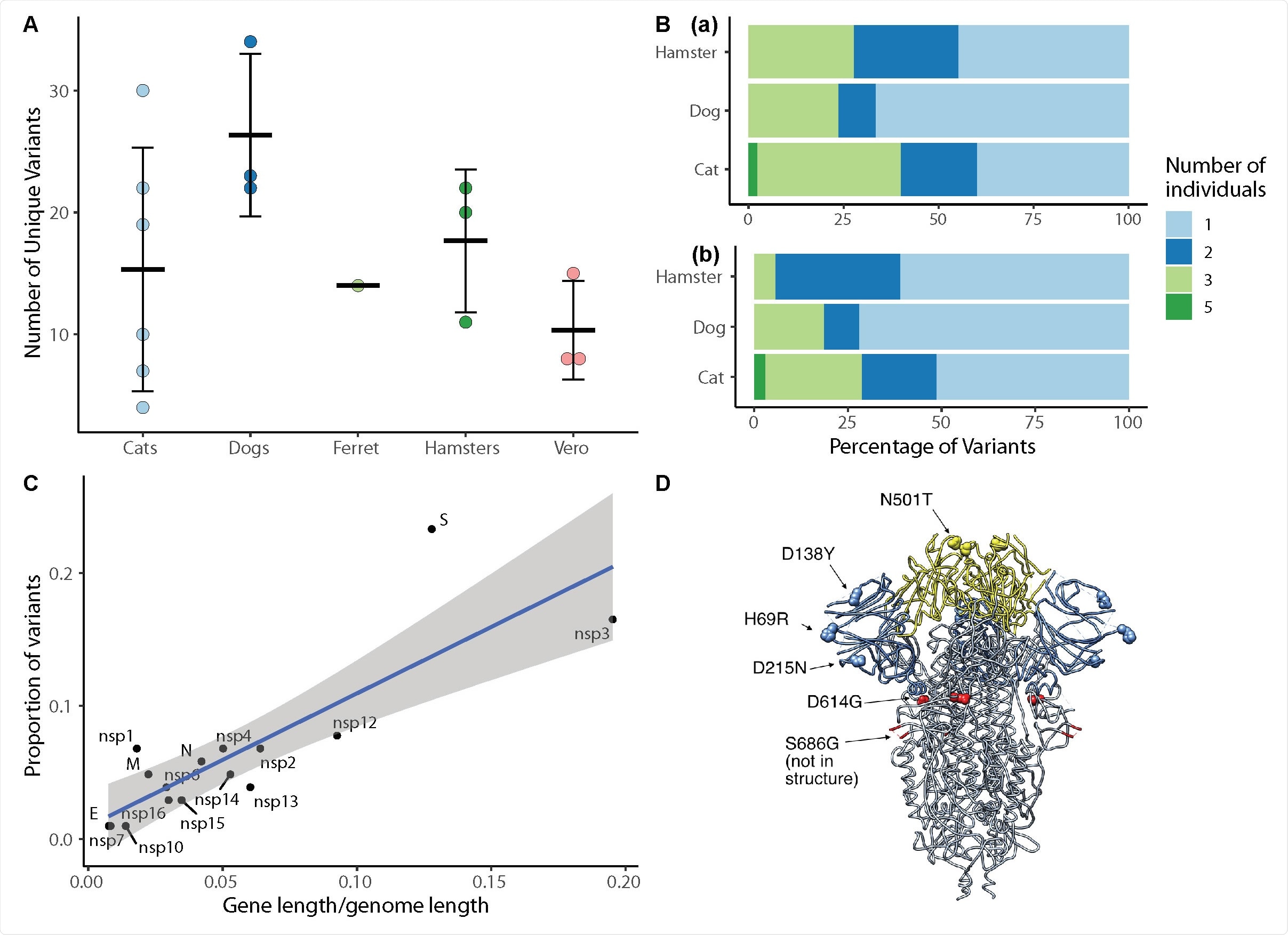
SARS-CoV-2 viral evolution differs across species, gene regions, and individuals. (A) Each point indicates the number of unique variants detected at ≥ 3% frequency in SARS-CoV-2 genomes recovered from individual animals. There is no significant difference in the number of unique variants detected in different species (ANOVA, p=0.22). (B) Analysis of variant distribution within species reveals that the majority of variants were detected in just one individual within each species. Subplot (a) shows the distribution for all variants, while (b) illustrates only variants not occurring in the P3 inoculum at >3%. (C) Variants are distributed across viral genes in relation to each gene’s length as a proportion of the entire genome length (linear model, R2=0.69, p<0.0001). The spike protein contained a notably higher proportion of all variants in comparison to its share of genome length. Grey shading represents the 95% confidence interval for the slope of the regression line. (D) SARS-CoV-2 spike protein variant “residues of concern” are in N terminal domain (NTD), receptor binding domain (RBD) and 670 furin cleavage site. Blue indicates NTD, Yellow indicates RBD. The furin cleavage site and adjacent residue 686 are in the indicated loop, which was not resolved in this structure.
The team found five chances in nsp12, S, N, and M genes near fixation in the inoculum. Still, it returned to wild-type sequences in the ribonucleic acid (RNA) recovered from the animals about one to three days after exposure.
The researchers detected 14 emergency variants in viruses recovered in the animals, including substitutions at spike positions N501, H69, and D614. The speed of in vitro and in vivo SARS-CoV-02 selection shows residues with functional importance during host-switching.
“Pathogens are under strong selective pressure to propagate in the host environment, while host defenses are aligned to prevent pathogen replication,” the team explained.
“This host-pathogen arms race results in varied outcomes that can lead to increases or decreases in virulence and transmission,” they added.
Hence, the study findings illustrate the potential for spillback reservoir hosts to hasten evolution. Further, the results highlight the plasticity of viral adaptation in animal models.
The findings may guide scientists to prevent future outbreaks that may emerge in animals and jump to humans.
To date, the coronavirus disease continues to ravage across the globe. The United States remains the country with the highest case toll, reaching over 29 million, followed by India, with over 11.24 million cases, and Brazil, with over11.12 million cases.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Bashor, L., Gagne, R., Lauth, A. et al. (2021). SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. bioRxiv., https://doi.org/10.1101/2021.03.05.434135, https://www.biorxiv.org/content/10.1101/2021.03.05.434135v2
- Peer reviewed and published scientific report.
Bashor, Laura, Roderick B. Gagne, Angela M. Bosco-Lauth, Richard A. Bowen, Mark Stenglein, and Sue VandeWoude. 2021. “SARS-CoV-2 Evolution in Animals Suggests Mechanisms for Rapid Variant Selection.” Proceedings of the National Academy of Sciences 118 (44). https://doi.org/10.1073/pnas.2105253118. https://www.pnas.org/doi/full/10.1073/pnas.2105253118.