An international team of researchers has found that glycosylation patterns are conserved across the native spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19) – and the recombinant spike proteins made in laboratories for use in vaccines.
The spike protein, which SARS-CoV-2 uses to bind to and infect cells, is the main target of neutralizing antibodies following natural infection with the virus or vaccination.
A central tenet in vaccine design is the presentation of native-like antigens that will induce protective immunity.
"The abundance of N-linked glycans across the SARS-CoV-2 spike protein is a potential source of heterogeneity between the many different vaccine candidates under investigation," says Max Crispin from the University of Southampton in the UK and colleagues.
Now, the team has conducted a study exploring this potential variability across five different recombinant spike proteins and the native spike using liquid chromatography-mass spectrometry (LC-MS) and molecular dynamics (MD) simulations.
The analysis revealed that glycosylation patterns were conserved across all spike proteins investigated, suggesting that the recombinant spike protein glycosylation likely mimics that of the native protein.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
Vaccine design has mainly focused on the spike protein
Since the COVID-19 outbreak first began in Wuhan, China, in late December 2019, researchers have developed an unprecedented array of vaccine candidates against the causative agent SARS-CoV-2.
All approaches aim to deliver immunogenic features of the virus, with the viral spike protein having emerged as the main focus owing to the robust antibody response it has been shown to generate.
Encouragingly, antibody neutralization has been shown to occur, despite the extensive array of N-linked glycans distributed across the spike protein. However, glycosylation has still become an important parameter in vaccine development as the glycosylation processing state can reveal the extent to which immunogens are recapitulating the native viral architecture.
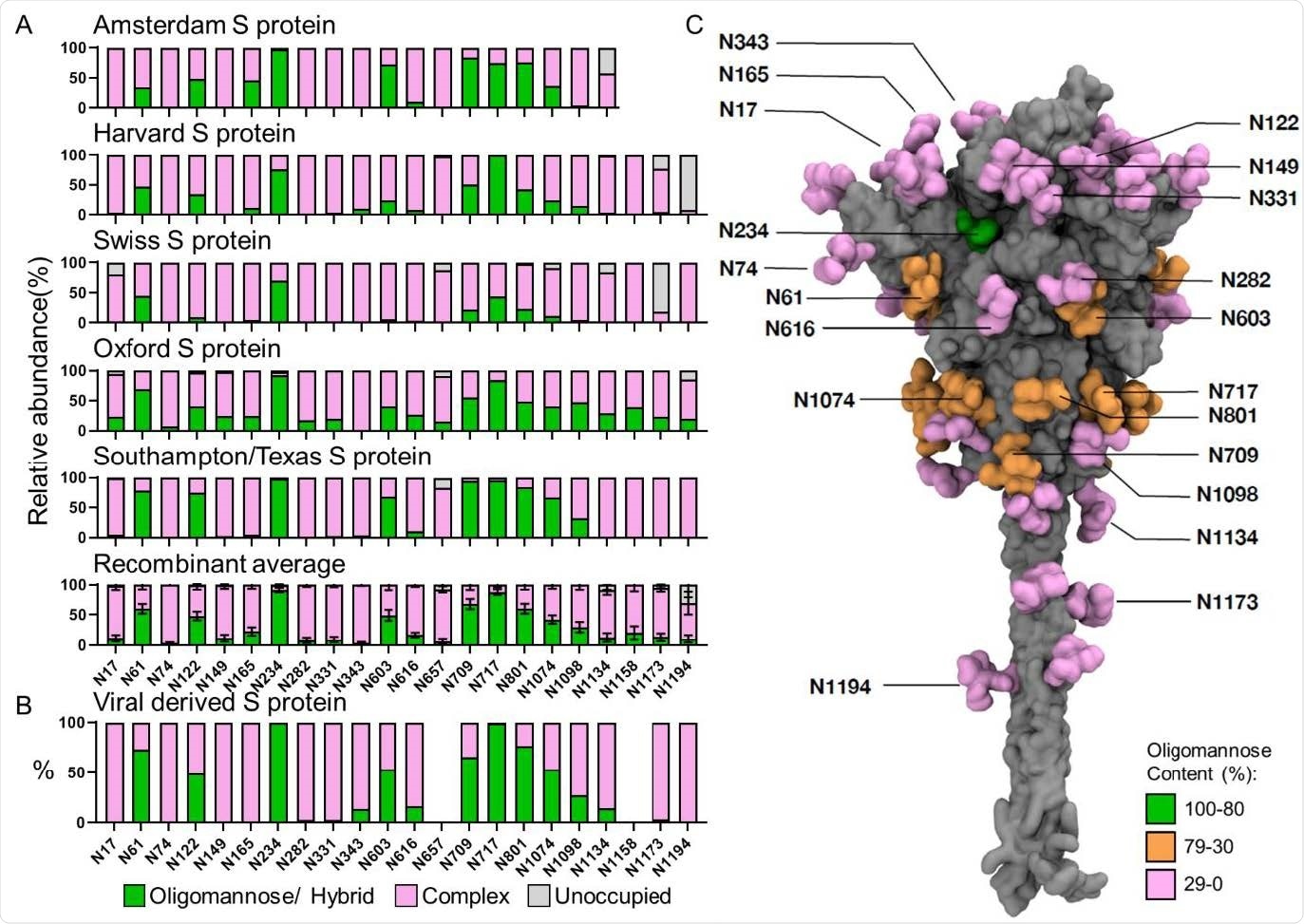
The site-specific glycosylation of recombinant and viral derived S protein from multiple laboratories. (A) Site-specific glycan analysis of recombinant S protein expressed and purified at different locations. The bar charts represent the relative proportion of glycoforms present at each site, including the proportion of PNGS that were not modified by an N-linked glycan. The proportion of oligomannose- and hybrid-type glycans are colored green. Processed complex-type glycans are colored pink and the proportion of unoccupied sites are colored grey. The institution which provided the S protein for analysis is listed above each chart. The Texas/Southampton data is reproduced from Chawla et al. (unpublished data). The average compositions of recombinant S protein were calculated using all samples. Bars represent the mean +/- standard error of the mean of all recombinant samples analyzed. (B) The viral-derived site-specific analysis was obtained from data acquired by Yao et al. and categorized in the same manner as above33. Data for sites N657 and N1158 could not be obtained and are not represented. (C) Full-length model displaying the site-specific oligomannose glycosylation of viral derived S protein; description of how this model was generated can be found in Materials and Methods. Both protein and glycans are shown in surface representation; the former colored in grey and the latter coloured based on oligomannose content (green for 80-100%, orange for 30-80% and pink for 0-30%).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The concerns surrounding glycosylation
The SARS-CoV-2 spike protein contains an abundance of N-linked glycosylation sequons that direct the attachment of host glycans required for protein folding and stabilization of the protein fold.
Although the spike protein has been an important focus in vaccine design, mechanisms of delivery have differed considerably.
Immunogen glycosylation can be influenced by factors specific to the particular manufacturing process, such as the cell type or cell culture conditions. The immunogen's design can also have a substantial impact, adversely affecting the presentation of native-like glycosylation.
"However, the success of a broad range of different vaccine platforms exhibiting different spike protein glycosylation indicates that native-like glycosylation is not a prerequisite for a successful vaccine," says Crispin and colleagues.
Nevertheless, understanding spike protein glycosylation will help standardize the materials used in different vaccine studies and help define the impact of this extensive feature of the protein surface, adds the team.
"It is important for therapeutic and vaccine design to understand whether similar glycoforms arise across different protein expression platforms from different sources to ensure the antigenic surface of the spike protein remains consistent when used as an immunogen or in serological assays."
What did the researchers do?
The researchers integrated LC-MS and MD simulations to investigate the glycosylation features across recombinant SARS-CoV-2 spike proteins from five different laboratories.
They then assessed the degree to which these recombinant proteins reproduced the glycosylation of SARS-CoV-2-derived spike protein produced from cultured Vero cells.
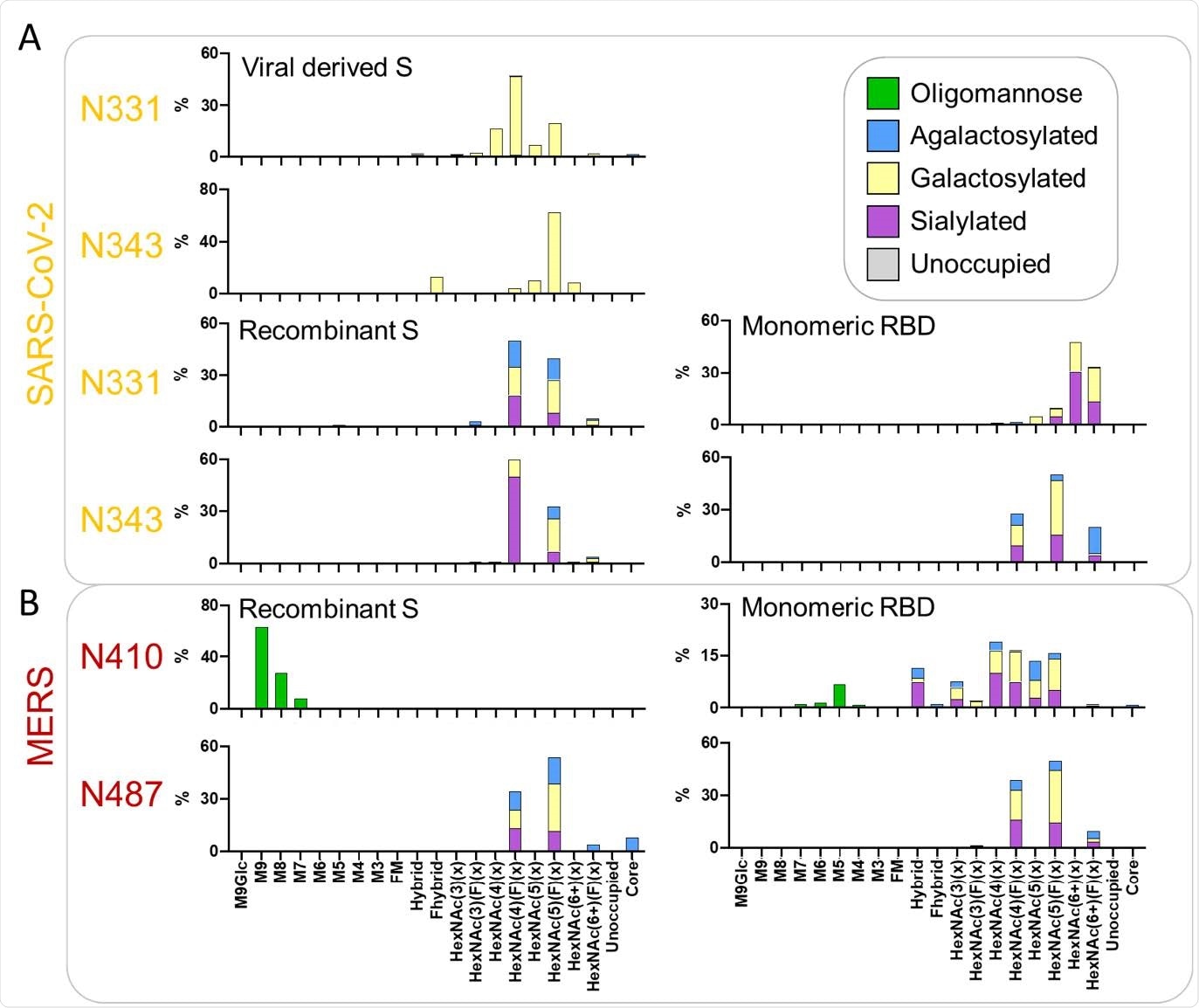
Comparative analysis of the glycosylation of the two PNGS on the RBD for viral derived S protein, recombinant S protein and monomeric RBD. (A) Detailed site-specific glycan compositions of the two sites located in the RBD of SARS-CoV-2. Recombinant S protein data is reproduced from Chawla et al. (unpublished) and data for the viral S protein RBD sites was obtained from Yao et al.33. Site-specific glycan data is presented as outlined in Figure 2. (B) Site-specific compositions for N-glycan sites located in the RBD of MERS-CoV when expressed as part of a soluble recombinant S protein compared to RBD-only. Data for the MERS-CoV S protein were obtained from a previous study.
What did they find?
Glycosylation patterns were conserved across all proteins investigated.
Distinct degrees of accessibility between different glycan sites across the spike protein broadly correlated with their processing states.
"Our results reveal the conserved structural N-glycan sites in spike protein when compared with native viral spike, which drives similarity in glycosylation amongst spike protein from disparate sources and manufacture methods," writes the team.
"This suggests that recombinant spike-based SARS-CoV-2 immunogen glycosylation reproducibly recapitulates signatures of viral glycosylation."
What are the study implications?
Glycosylation of recombinant spike proteins used in vaccinations likely mimics the patterns occurring in natural infection and will remain antigenically comparable, say the researchers.
However, continued understanding of spike protein glycosylation will further facilitate vaccine research during the global COVID-19 response, they advise.
"The reproducibility of spike protein glycosylation from many different sources is of significant benefit for immunogen design, serology testing and drug discovery and will mean that the glycans of SARS-CoV-2 are unlikely to provide a barrier to combatting the COVID-19 pandemic," concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Crispin M, et al. Site-specific steric control of SARS-CoV-2 spike glycosylation. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.08.433764, https://www.biorxiv.org/content/10.1101/2021.03.08.433764v1
- Peer reviewed and published scientific report.
Allen, Joel D., Himanshi Chawla, Firdaus Samsudin, Lorena Zuzic, Aishwary Tukaram Shivgan, Yasunori Watanabe, Wan-ting He, et al. 2021. “Site-Specific Steric Control of SARS-CoV-2 Spike Glycosylation.” Biochemistry 60 (27): 2153–69. https://doi.org/10.1021/acs.biochem.1c00279. https://pubs.acs.org/doi/10.1021/acs.biochem.1c00279.