Researchers in the United States have found that protective antibodies generated against the original strain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)– the agent that causes coronavirus disease 2019 (COVID-19) – also protect against viral variants.
Monoclonal antibodies derived from memory B cell reserves in convalescent individuals competed to bind the spike protein of both the original SARS-CoV-2 strain and variant strains. The spike protein is the main surface structure the virus uses to bind to and infect host cells.
Duane Wesemann from Harvard Medical School in Boston and colleagues say the study maps a crucial component of the long-term immune response to SARS-CoV-2 infection.
“The results furnish a global atlas of the spike-specific memory B cell repertoire and illustrate properties conferring robustness against emerging SARS-CoV-2 variants,” they write.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
.jpg)
Study: Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Image Credit: NIAID
Understanding immune recognition of new variants is crucial

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Since the COVID-19 outbreak first began in Wuhan, China, in late December 2019, researchers have raced to develop vaccines against the causative agent SARS-CoV-2. While several vaccines have received emergency use authorization in record time, new variants continue to emerge, threatening to evade immune responses.
“We need to understand immune recognition of SARS-CoV-2, especially as stored in B cell memory, to illuminate the requirements for broad protective immunity,” says Wesemann and the team.
Antibodies are both effector molecules and the antigen-receptor component of the B cell receptor (BCR). BCRs evolve enhanced pathogen binding through immunoglobulin gene somatic hypermutation and selection in the germinal centers of lymphoid tissue. This leads to the maturation of antibody affinity and the generation of both antibody-secreting plasma cells and memory B cells.
Antibodies wane over time
The spike protein is the main target of neutralizing antibodies following infection with SARS-CoV-2 or vaccination. Levels of these antibodies wane over time and can lose reactivity to emerging variants.
However, studies have shown that antibodies cloned from memory B cells target the spike in both redundant and unique ways, pointing to both cooperative and competitive recognition.
“Many of these antibodies have been identified and characterized; their positions within the distribution of practical cooperative recognition of SARS-CoV-2 spike within the human memory B cell repertoire have not,” write the researchers.
“Moreover, the recognition reach of memory B cells induced by one SARS-CoV-2 strain toward evolving strains across the major epitopic regions has not yet been defined.”
What did the researchers do?
The team tested 152 memory B cell receptor-encoded monoclonal antibodies (mAbs) from convalescent plasma for binding of the SARS-CoV-2 spike protein.
The blood samples used had been drawn from 19 individuals who had developed COVID-19 symptoms between the 3rd March and 1st April, 2020. Therefore, these individuals would have generated immune responses against viral strains that were circulating long before the UK and South African variants were first reported in December 2020.
“This set of BCR sequences and corresponding mAbs thus represents responses to a relatively homogeneous infectious virus and provides a valuable tool for examining the degree to which these antibodies retain recognition of emerging variants and for studying the extent to which loss of neutralizing titer correlates with loss of longer-term protection,” says Wesemann and colleagues.
Clusters of mAbs were mapped onto the spike protein by including previously characterized antibodies, as well as new structures determined by cryogenic electron microscopy.
What did the study find?
The team identified 7 recurrently targeted mAb competition groups against epitopes on the spike protein – three groups with epitopes on the receptor-binding domain (RBD-1 to RBD-3), two with epitopes on the N-terminal domain (NTD-1 and NTD-2), and two with epitopes on subunit 2 (S2-1 and S2-2).
“We show that these groups represent the major practical antibody footprints, with rare antibodies outside them,” write the researchers.
Antibodies mapped to RBD-2 and NTD-1 were the most potent neutralizers, while those mapped to S2-1 exhibited the greatest recognition breadth across different strains.
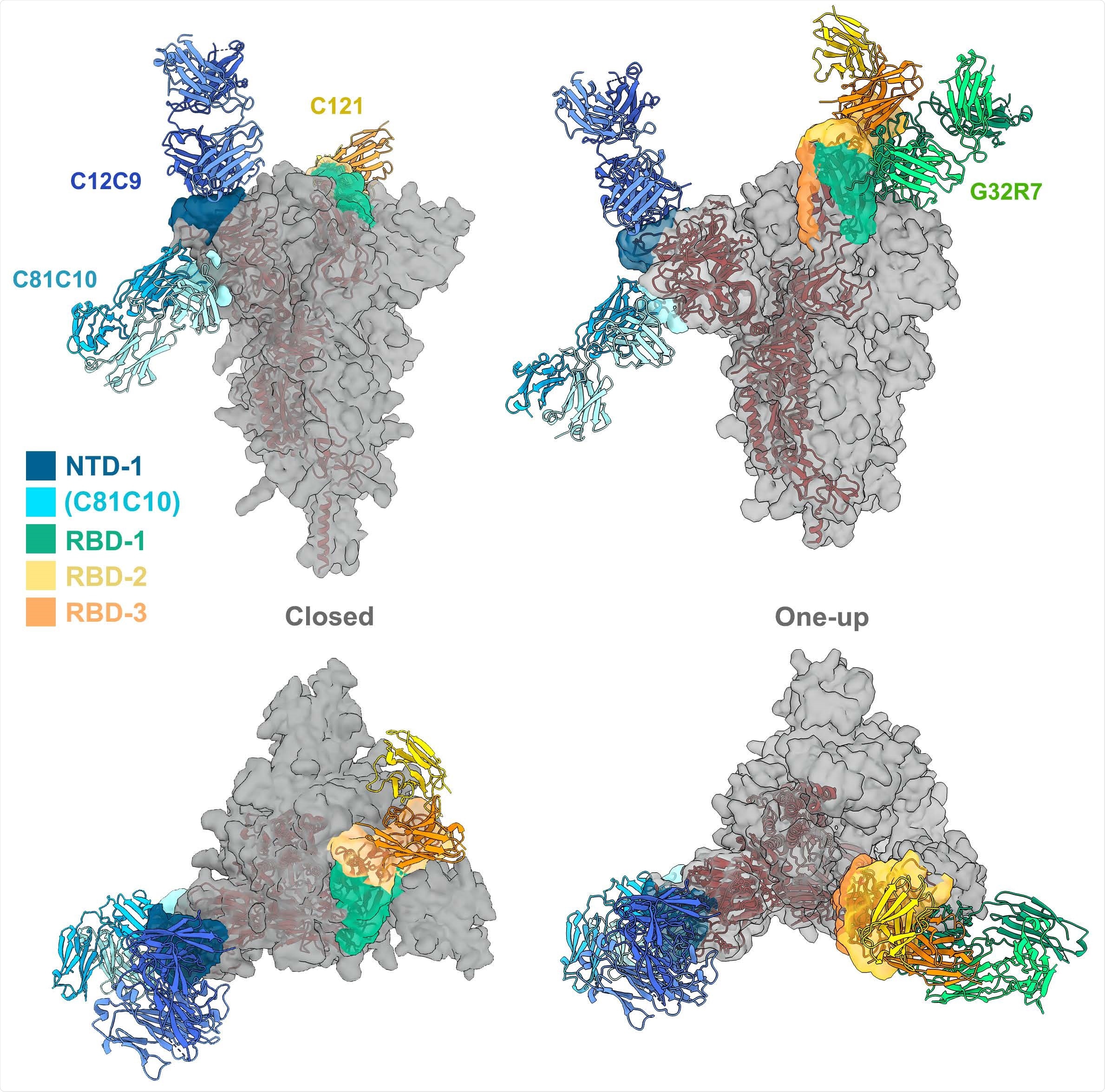
Ab contact regions. Surface regions of the SARS-CoV-2 spike protein trimer contacted by antibodies in four of the seven principal clusters, according to the color scheme shown (taken from the color scheme in Fig. 2), with a representative Fab for all except RBD-3. The C81C10 Fab defines an epitope just outside the margin of NTD-1, but it does not compete with any antibodies in RBD-2. The RBD-2 Fv shown is that of C121 (PDB ID: 7K8X: Barnes et al, 2020), which fits most closely, of the many published RBD-2 antibodies, into our low-resolution map for C12A2. Left: views normal to and along threefold axis of the closed, all-RBD-down conformation; right: similar views of the one-RBDup conformation. C121 (RBD-2) can bind both RBD down and RBD up; G32R7 (RBD-1) binds only the "up" conformation of the RBD. The epitopes of the several published RBD-3 antibodies are partly occluded in both closed and open conformations of the RBD; none are shown here as cartoons. A cartoon of the polypeptide chain of a single subunit (dark red) is shown within the surface contour for a spike trimer (gray).
When the team tested the antibodies against emerging SARS-CoV-2 variants, antibodies in one of the RBD and one of the NTD clusters were significantly affected, particularly by the South Africa strain.
The mutations present in these variants influenced the binding affinity of antibodies within a competition group differently, suggesting that the depth of otherwise redundant mAbs confers protection.
What are the implications of the study?
“Our data suggest an additional mechanism for preventing viral escape: competing antibodies may help retain recognition of a rapidly evolving antigen by their differential sensitivity to specific mutations,” says Wesemann and the team.
The researchers say the potential dynamic reach of otherwise redundant mAb recognition may confer a selective advantage to immune mechanisms that yield multiple competing antibodies to critical epitopes, as those that retain adequate affinity can then re-activate, expand, and potentially undergo further affinity maturation.
“The emergence of strains that may have gained selective advantage by escape from neutralization emphasizes the importance of determining whether the level of retained affinity for the spike protein by some antibodies in the immunodominant clusters influences protection from clinical disease,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tong P, et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.10.434840, https://www.biorxiv.org/content/10.1101/2021.03.10.434840v1
- Peer reviewed and published scientific report.
Tong, Pei, Avneesh Gautam, Ian W. Windsor, Meghan Travers, Yuezhou Chen, Nicholas Garcia, Noah B. Whiteman, et al. 2021. “Memory B Cell Repertoire for Recognition of Evolving SARS-CoV-2 Spike.” Cell 184 (19): 4969-4980.e15. https://doi.org/10.1016/j.cell.2021.07.025. https://www.cell.com/cell/fulltext/S0092-8674(21)00884-9.