A recent environmental study from the United Kingdom, currently available on the bioRxiv* preprint server, shows that certain common surfaces could represent an infectious risk if contaminated with high concentrations of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – albeit the level of viable viral particles is likely to be low.
The global pandemic of coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, was successful due to viral transmission via a number of different routes, including human contact with contaminated surfaces. People usually get contaminated either as a result of virus-containing droplets or by touch.
Consequently, molecular analytical approaches were used by many researchers to detect SARS-CoV-2 genetic material on various surfaces; however, a viable virus was rarely found. This means that the risk of infection from virus-contaminated surfaces is rather challenging to predict, invoking further studies to enhance our understanding of its survivability.
This new paper by UK researchers (led by Dr. Susan Elizabeth Paton from the Public Health England, National Infection Service in Wiltshire) aimed to appraise the viability of SARS-CoV-2 over a certain period after being dried onto a range of materials, but also to compare the viability of the virus to RNA copies recovered and observe its potential concentration dependence.
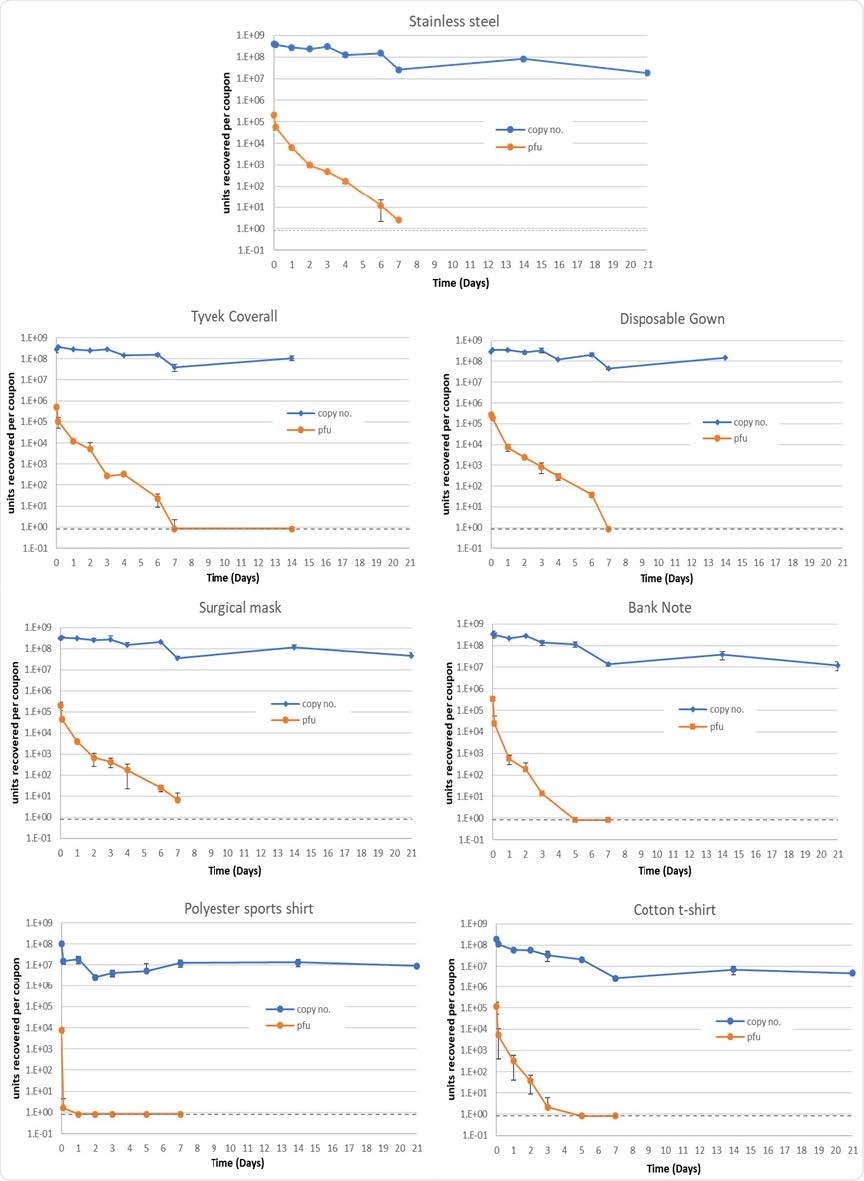
Mean quantities of viable virus recovered (pfu/coupon, orange) and viral RNA detected (genome copy number/coupon, blue), for 7 materials assessed. Error bars represent the standard deviation from three replicates. Grey dashed line represents the limit of detection of the plaque assay for the combined assays from the triplicate coupons (0.8 pfu/ml). For Tyvek coverall and disposable gown, 21-day coupons were not processed.
Inoculating common materials
In this study, the researchers decided to use surfaces representative of non-porous hand-touch sites (such as stainless steel) and banknotes (i.e., English polymer 10 GBP notes) and personal protective equipment items that are pervasive in the health care setting and the wider environment, such as layered surgical masks, disposable plastic gowns, and coveralls. This was supplemented by materials that represented clothing items, such as polyester sports shirts and cotton T-shirts. Coupons sized 1x1 cm of each material have been prepared.
The SARS-CoV-2 strain was initially isolated using Vero E6 cells (derived from the kidney of an African green monkey), and viral aliquots were defrosted to room temperature immediately before the inoculation of materials. The virus was recovered from the coupons, and a plaque assay was then conducted. RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR) analysis have also been pursued.
When data analysis is concerned, each time point for every material coupon used had three biological replicates (i.e., individual coupons) and two technical replicates (i.e., plaque assay performed in duplicate).
Optimal stability on non-porous hydrophobic surfaces
This study demonstrated that the UK SARS-CoV-2 isolate could remain viable on hydrophobic surfaces for up to seven days. Its recoverable viability on hydrophilic surfaces is basically reduced to three days at relative humidity ambient temperature.
More specifically, viable virus persisted the longest on stainless steel and surgical mask material (more than 100 hours for 99.9% reduction in SARS-CoV-2 viability for both materials), while the fastest drop was observed on a polyester shirt (99.9% reduction in 2.5 hours), followed by cotton (99.9% reduction in 72 hours).
In any case, SARS-CoV-2 was most stable on non-porous hydrophobic surfaces, and viral RNA was very stable when dried on surfaces – with only one log reduction in recovery over three weeks. Still, it was shown that SARS-CoV-2 viability decreased much more rapidly, and such decay rate was independent of starting concentration.
Genetic material vs. viable virus
The study concludes that expected levels of viable SARS-CoV-2 environmental surface contamination lead to undetectable levels within two days. In other words, when viral genetic material (RNA) is detected on surfaces, it does not directly suggest the presence of a viable virus.
"This has implications for interpretation of surface sampling results using RT-PCR to determine the possibility of viable virus from a surface," caution study authors in this bioRxiv paper. "Unless sampled immediately after contamination, it is difficult to align RNA copy numbers to the quantity of viable virus on a surface", they add.
Future studies may address this current limitation, where the copy number is obtained for much lower concentrations of SARS-CoV-2 present on surfaces. Consequently, this will help identify whether the persistence of RNA is independent of viral concentration and elucidates the exact relationship to viable virus recovery.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Paton, S.E., et al. (2021). Persistence of SARS-CoV-2 virus and viral RNA on hydrophobic and hydrophilic surfaces and investigating contamination concentration. bioRxiv. https://doi.org/10.1101/2021.03.11.435056, https://www.biorxiv.org/content/10.1101/2021.03.11.435056v1
- Peer reviewed and published scientific report.
Paton, Susan, Antony Spencer, Isobel Garratt, Katy-Anne Thompson, Ikshitaa Dinesh, Paz Aranega-Bou, David Stevenson, et al. 2021. “Persistence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Virus and Viral RNA in Relation to Surface Type and Contamination Concentration.” Applied and Environmental Microbiology 87 (14): e0052621. https://doi.org/10.1128/AEM.00526-21. https://journals.asm.org/doi/10.1128/AEM.00526-21.