The machinery for viral replication and transcription is translated from particular sections of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome, open reading frames 1a and 1b (ORF1a and ORF1b). Sixteen non-structural proteins (NSP) are produced upon translation. Of these, some appear to be highly conserved across coronavirus lineages due to the vital and essential role these proteins play in replication, making the occurrence of successful and distinct mutants rare. In a research paper recently uploaded to the preprint server bioRxiv* by Newman et al. (March 15th, 2021) the structure and role of one such protein: NSP13, is investigated, with two appealingly druggable targets being identified, possibly opening a new route towards a novel SARS-CoV-2 antiviral agent.
What is NSP13?
NSP13 is a 67 kDa helicase protein, helicases being a class of enzymes that separate nucleic acid strands and thus are directly involved in many cellular processes such as replication, transcription, and translation. NSP13 belongs to the helicase superfamily 1B, this further categorization indicating that the protein does not form a ring structure, as is the case with all known RNA helicases in eukaryotes, though some ring-forming helicases have been found in viruses, and that the enzyme performs translocation in the 5’-3’ direction.
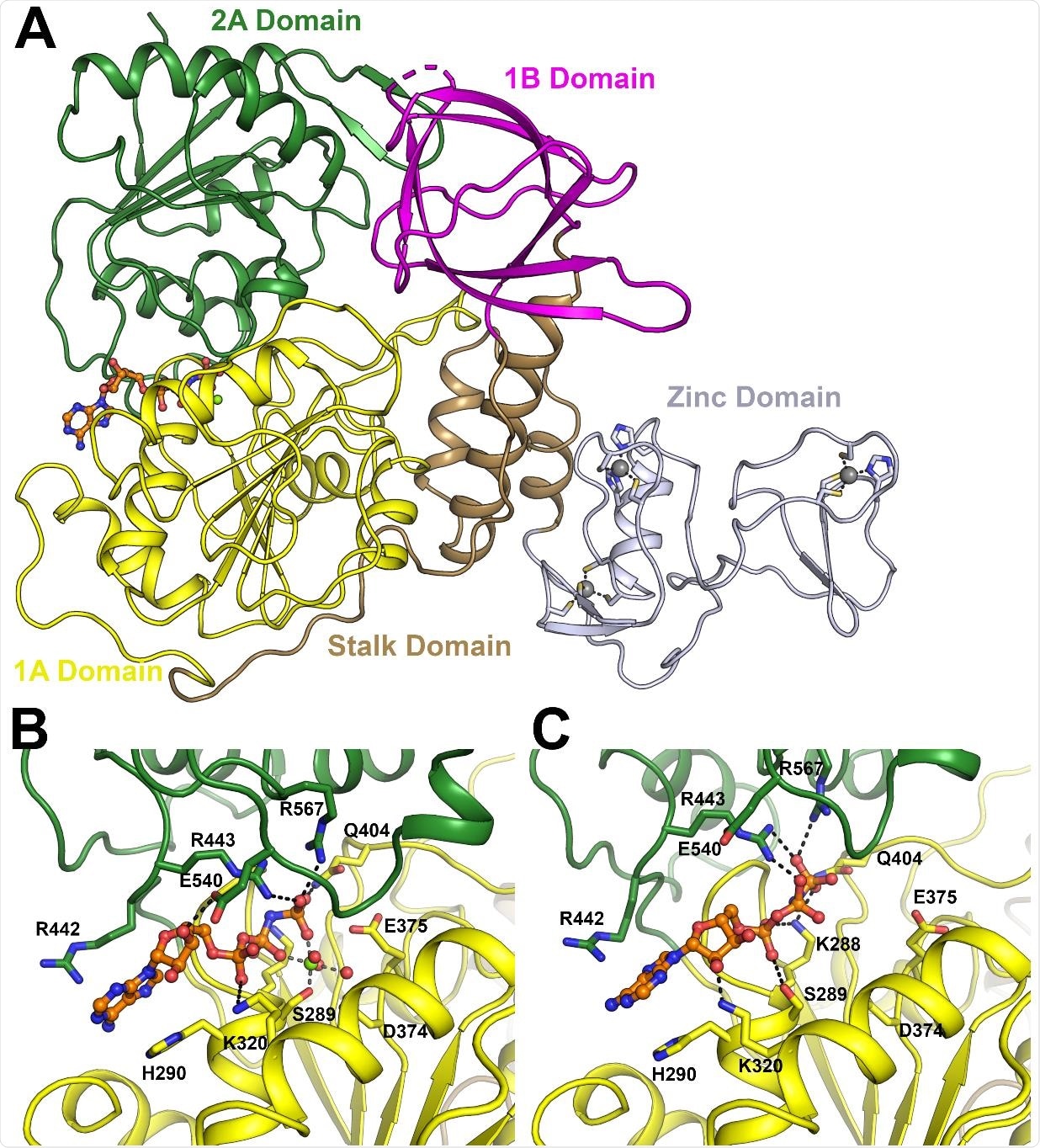
Overall structure of SARS-CoV-2 NSP13. (A) Structure overview with domains labelled and colored individually (the same color scheme is used throughout). The AMP-PNP nucleotide is shown in stick format in the nucleotide binding site between the 1A and 2A domains. (B) Close up view of the nucleotide binding mode for the AMP-PNP Mg2+ complex (mode A), with interacting residues labeled and shown in the stick format. (C) Close up view of the nucleotide binding form the AMP-PNP complex (mode B), viewed from the same orientation as panel B.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
NSP13 contains 5 domains: an N-terminal zinc-binding domain, a helical stalk, a beta-barrel, and two binding domains that make contact with the nucleotides. The protein is also structurally very similar in both MERS-CoV and SARS-CoV-1, differing by only a single amino acid in the case of the latter, and so has been proposed as a promising target for antiviral drugs. Some preliminary drug development has already taken place towards drugging this particular protein target, given its strongly conserved presence even in more distantly related viruses still. However, the precise structure of this protein has not yet been entirely elucidated for SARS-CoV-2. X-ray crystallographic structures are the gold standard in determining protein conformation in various in situ modes, accounting for when bound with target nucleotides and when part of the transcription supercluster of proteins. The group uncovered two likely binding pockets in this manner, and further performed a fragment screening of the protein to test the ligandability, providing a starting structure upon which the rational design of drugs can be based.
The crystal structure of NSP13
Three conformations of the proteins were examined: the APO form, when free and not bound with the associated enzyme cofactor proteins; the phosphate bound form, which is active; and the nucleotide bound form, representative of when engaged in enzymatic activity. In each case, the chains were noted to be similar to as described previously for NSP13 in related organisms. However, while the chains were of similar sequence, the overall conformational structure was found to differ in some regions by as much as 3 Å, attributed to the flexible zinc-binding domain and nucleotide-binding domains.
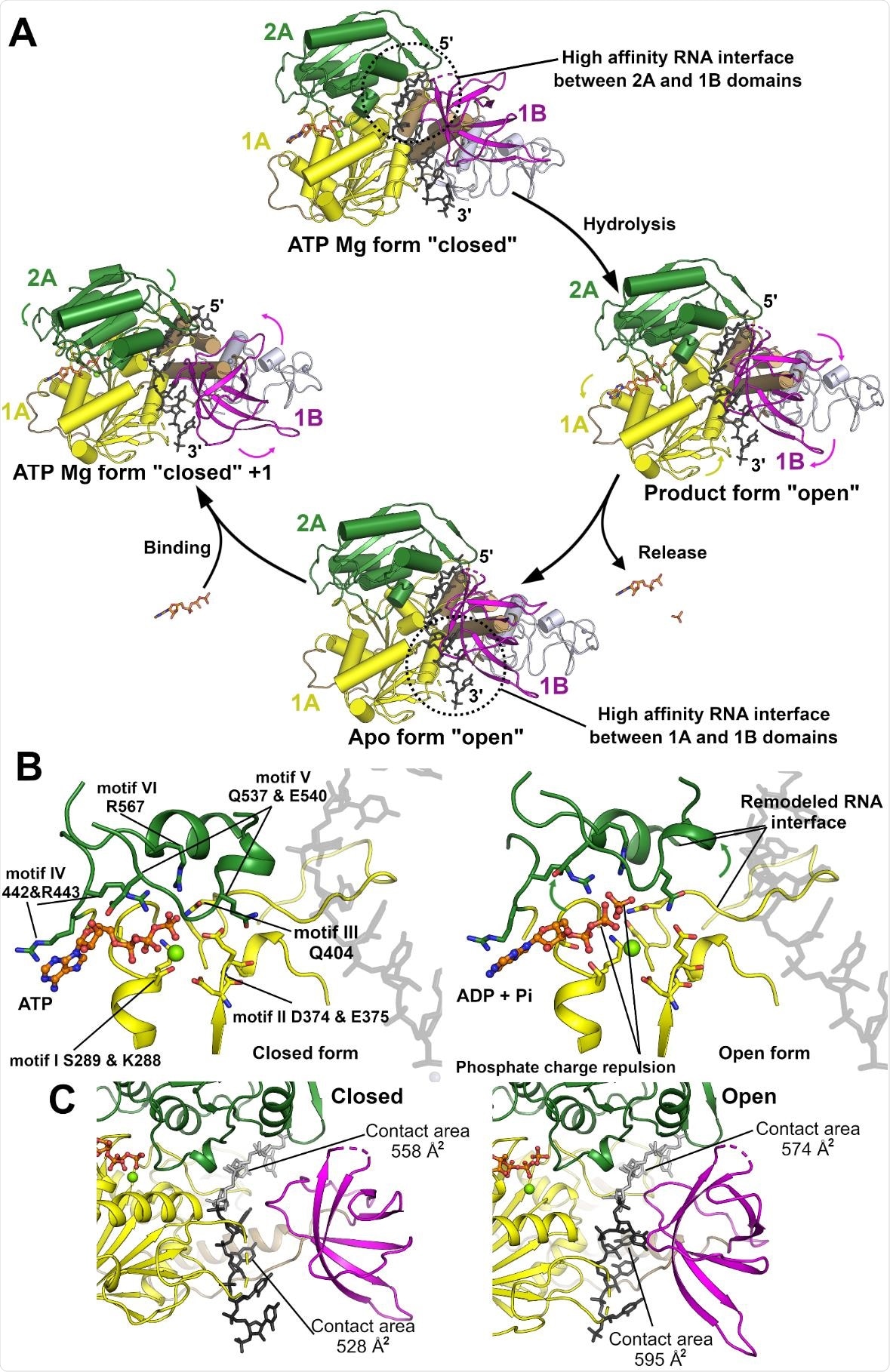
Model for the NSP13 5’ to 3’ translocation mechanism. (A) Proposed translocation mechanism for NSP13 based on the transition from the closed (pre hydrolysis) to open (Product and APO) forms. The transitions are initiated by the binding, hydrolysis and release of ATP which triggers the conformational changes and remodels the RNA interface. (B) Close up view of the active site with ATP in the closed conformation (left) and ADP and Pi in the open conformation (right). Hydrolysis as subsequent charge repulsion could trigger the opening of the cleft between the two domains with conserved motifs on the 2A domain primarily contacting the product phosphate whilst the ADP product interacts with the 1A domain. Several of the phosphate interacting motifs are proximal to regions of the RNA binding interface indicating the possibility of modulation based on hydrolysis status. (C) Remodeling of the RNA interface based on the position adopted by the 1B domain. The closed conformation is shown on the left and the open on the right. The contact areas for the 5’ and 3’ RNA regions (depicted in grey and black) is shown.
Specific changes in the nucleotide-binding sites could be observed as the protein transitioned between open and closed states, to which the group ascribe the translocation mechanism of the protein along the nucleotide chain. When in the closed conformation, RNA binds with the protein preferentially through one of the two binding domains, with the weaker bond towards the 3’ end. ATP hydrolysis then triggers the conformational change to the open state, reversing the affinities and shifting the protein along the chain by one nucleotide.
Crystallographic fragment screening
The phosphate-bound form of NSP13 was crystallized, and 648 crystals individually soaked in a library of chemical fragments. X-ray analysis was then performed, and any bound ligands were identified. 65 fragments were identified to bind to various sites of the protein, 15 of which overlapped the ATP ribose moiety. Several others were located close to the RNA/DNA binding pocket and would be expected to block interaction with the protein. A small number of additional ligands were expected to bind the zinc domain of the protein with the stalk domain, disallowing the conformational changes necessary to engage in nucleotide bonding.
Further computational analysis of target sites was performed, in particular, noting the 5’-RNA binding pocket as eminently druggable due to the hydrophobic environment, good accessibility when the protein is in the open conformation, and high conservation across other coronaviruses.
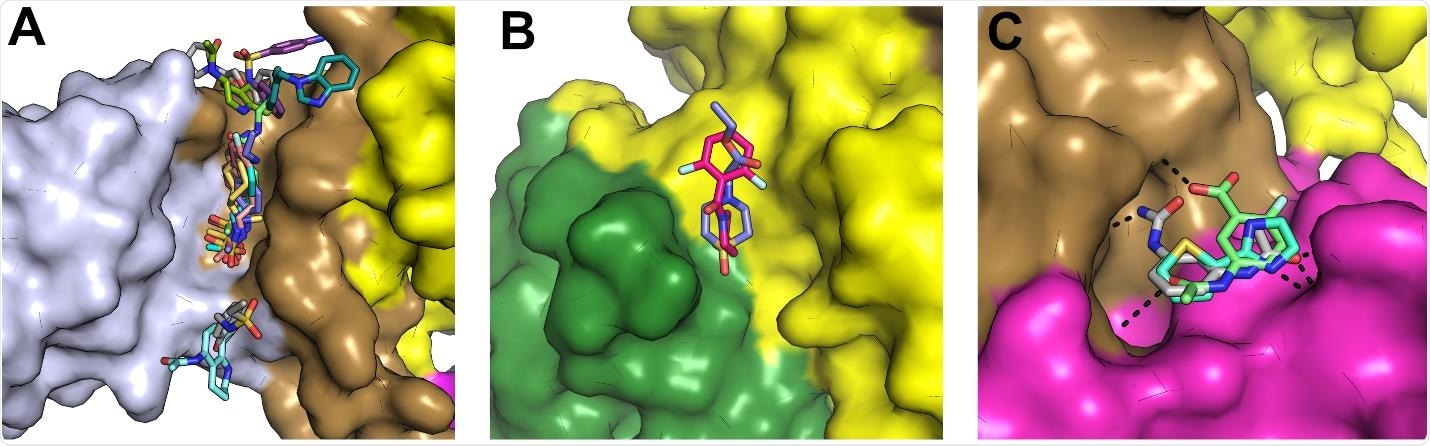
Fragments bound on the interfaces between domains that appear to move as part of the catalytic cycle and thus may be starting points for allosteric inhibitor design. (A) Surface view of a prominent fragment binding site which bound to 11 fragments in a cleft between the Zinc and Stalk domains with the domains colored individually as for the color scheme in Fig1. (B) Two fragments were observed to bind in a cleft between the 1A and 2A domains approximately opposite the hinge. (C) Three fragments were bound in a shallow pocket between the Stalk and 1B domain (also close to the RNA interface) and make polar contacts to both regions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Structure, Mechanism and Crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Joseph A Newman, Alice Douangamath, Setayesh Yazdani, Yuliana Yosaatmadja, Anthony Aimon, José Brandão-Neto, Louise Dunnett, Tyler Gorrie-stone, Rachel Skyner, Daren Fearon, Matthieu Schapira, Frank von Delft, Opher Gileadi. bioRxiv. 2021.03.15.435326; doi: https://doi.org/10.1101/2021.03.15.435326, https://www.biorxiv.org/content/10.1101/2021.03.15.435326v1
- Peer reviewed and published scientific report.
Newman, Joseph A., Alice Douangamath, Setayesh Yadzani, Yuliana Yosaatmadja, Antony Aimon, José Brandão-Neto, Louise Dunnett, et al. 2021. “Structure, Mechanism and Crystallographic Fragment Screening of the SARS-CoV-2 NSP13 Helicase.” Nature Communications 12 (1): 4848. https://doi.org/10.1038/s41467-021-25166-6. https://www.nature.com/articles/s41467-021-25166-6.