New preclinical research led by Wensheng Wei from Peking University in China has proposed the development of a circular RNA vaccine. Their findings showed the circular RNA vaccine creates neutralizing antibodies and strong T-cell responses against the receptor-binding domain of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein.
The circular RNA vaccine also effectively neutralized the mutated receptor binding domain found in the B.1.351 variant. The B.1.351 variant was initially discovered in South Africa and could evade the immune system.
Currently approved messenger RNA (mRNA) vaccines from Moderna and Pfizer-BioNTech were developed before the variants emerged, so information is limited. The single-shot Johnson & Johnson vaccine was effective against the original strain in Wuhan, China, and the B.1.1.7 variant was first reported in South London last fall.
The results could help in dealing with the B.1351 variant and other variants of concern.
The authors write:
"Given that SARS-CoV-2 variants encoding E484K or N501Y or the K417N-E484K-N501Y evade certain neutralizing antibodies induced by mRNA vaccines, we anticipated that the effect of circRNA-encoded hACE2 decoy might not be affected by virus mutations."
The study "Circular RNA Vaccines against SARS-CoV-2 and Emerging Variants" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Development of the circular RNA vaccine
The team used a Group I ribozyme autocatalysis strategy to produce circular RNA encoding antigens specific to the receptor-binding domain of SARS-CoV-2 (circRNARBD). A signal peptide sequence was added to the receptor-binding domain's N-terminus because of its secretory expression of antigens.
The IRES element was positioned before the receptor-binding domain coding sequence to start translation. An IRES-SP-RBD-T4 sequence was added to the cyclic vector to produce circRNARBD.
Circular RNA model shows high protein expression and thermal stability
Their circular RNA model proved to be more resistant against RNAase R than linear RNA. When the purified circRNARBD was placed into HEK293T cells, they found many antigens — 50-fold more than linear RNARBD groups specific to the receptor-binding domain of SARS-CoV-2. These antigens prevented a SARS-CoV-2 pseudovirus infection.
They also showed high thermal stability. When stored at room temperature before transfection into HEK293T cells, circRNARBD continued to show expression two weeks after storage.
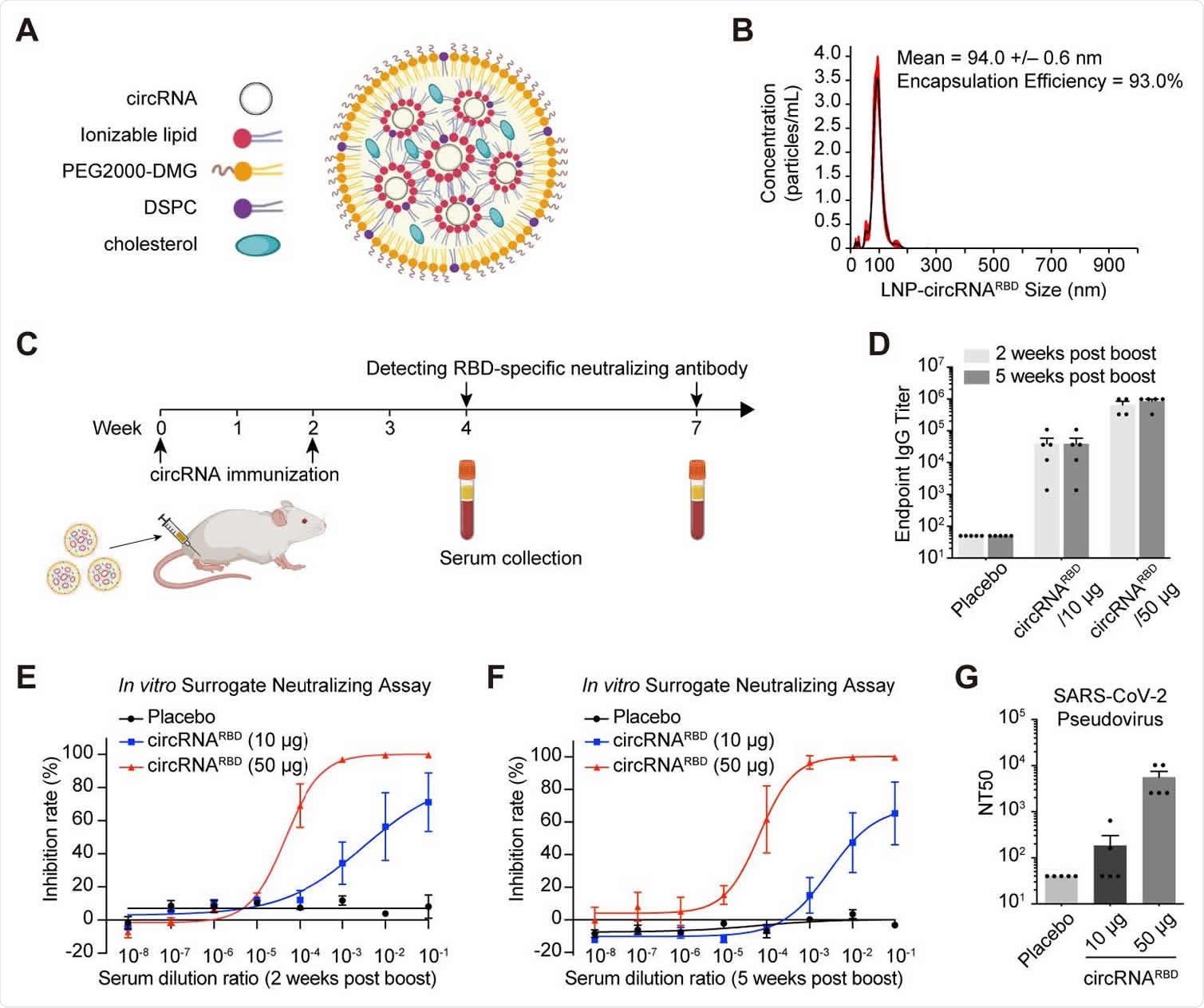
Humoral immune responses in mice immunized with SARS-CoV-2 circRNARBD vaccines. (A) Schematic representation of LNP-circRNA complex. (B) Representative of concentration-size graph of LNP-circRNARBD measured by dynamic light scattering method. (C) Schematic diagram of the LNP-circRNARB vaccination process in BALB/c mice and serum collection schedule for specific antibodies analysis. (D) Measuring the SARS-CoV-2 specific IgG antibody titer with ELISA. The data were shown as the mean ± S.E.M. (n = 4 or 5). (E) Sigmoidal curve diagram of the inhibition rate by sera of immunized mice with surrogate virus neutralization assay. Sera from circRNARBD (10 μg) and circRNARBD 260 (50 μg) immunized mice were collected at 2 weeks post the second dose. The data were shown as the mean ± S.E.M. (n = 4). (F) Sigmoidal curve diagram of the inhibition rate by sera of immunized mice with surrogate virus neutralization assay. Sera from circRNARBD (10 μg) and circRNARBD (50 μg) immunized mice were collected at 5 weeks post boost. The data were shown as the mean ± S.E.M. (n = 5). (G) The NT50 was calculated using lentivirus-based SARS-CoV-2 pseudovirus. The data were shown as the mean ± S.E.M. (n = 5).
Robust immune response in mice
Mice were intramuscularly injected with either 10 μg or 50 μg of experimental circRNARBD vaccine at a two-week interval. Immunity was measured two or five weeks after their booster shot.
The titers of immunoglobulin G (IgG) and antibodies were expressed in a dose-dependent manner and persisted two and five weeks after their booster shot. It also effectively neutralized a SARS-CoV-2 pseudovirus. The researchers suggest the circRNARBD vaccine creates an enduring immune response against SARS-CoV-2.
When evaluating CD4+ and CD8+ T cell immune responses after vaccination, the researchers found Th1-biased responses which produced interferon-γ (IFN-γ), tumor necrosis factor (TNF-α), and interleukin-2 (IL-2). However, changes in interleukin-4 (IL-4) were not observed. The results of the circular RNA vaccines stimulated a Th1 but not Th2 immune response.
There were also several cytokine-producing CD8+ detected in circRNARBD vaccinated mice. Interestingly, the team also found more robust immune responses in CD4+ and CD8+ effector memory T cells at 10 μg than 50 μg. Although, 50 μg induced higher potency of neutralizing antibodies in B cell responses.
Circular RNA vaccine is effective in neutralizing the B.1.351 variant
The team collected the serum of immunized mice 1 and 2 weeks after their booster shot. They found IgG titers specific for the spike protein's receptor binding domain with the 501.YV2 mutation.
The researchers next assessed mice's neutralizing activity with either the circRNARBD or circRNARBD-501Y.V2 vaccines against the D614G, B.1.1.7/501Y.V1, or B.1.351/501Y.V2 variants. The circRNARBD vaccine antibodies effectively neutralized all three viral strains with the highest activity against the D614G strain.
In contrast, the circRNARBD-501Y.V2 also neutralized all strains, with the highest neutralization activity against the corresponding variant, 501Y.V2.
"It's worth noting that both vaccines could neutralize all three strains albeit with variable efficacies. Nevertheless, the multivalent vaccines should have provided better protection for both native SARS-CoV-2 strain and its circulating variants," wrote the research team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Qu L, et al. Circular RNA Vaccines against SARS-CoV-2 and Emerging Variants. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.16.435594, https://www.biorxiv.org/content/10.1101/2021.03.16.435594v1
- Peer reviewed and published scientific report.
Qu, Liang, Zongyi Yi, Yong Shen, Liangru Lin, Feng Chen, Yiyuan Xu, Zeguang Wu, et al. 2022. “Circular RNA Vaccines against SARS-CoV-2 and Emerging Variants.” Cell, April. https://doi.org/10.1016/j.cell.2022.03.044. https://www.cell.com/cell/fulltext/S0092-8674(22)00394-4.