Mutations to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome are increasingly documented around the world, the minority of which are to the spike protein and receptor-binding domain (RBD) of the virus. However, this small number of mutations has been associated with the evasion of innate polyclonal antibodies developed by past SARS-CoV-2 infection in hosts, as most of these neutralizing antibodies target the RBD. In a paper recently uploaded to the preprint server bioRxiv*, Greaney et al. (March 18th, 2021) comprehensively map the RBD mutations that allow the virus to better evade capture against three selected neutralizing monoclonal antibodies and polyclonal antibodies isolated from the plasma of patients. The group identified a single class of antibodies that target a particular epitope on the RBD of SARS-CoV-2, suggesting that the human polyclonal response to SARS-CoV-2 infection is dominated by a worryingly narrow range of antibodies that target an epitope undergoing rapid mutation.
Which antibodies target SARS-CoV-2?
Antibodies that target the RBD of SARS-CoV-2 have been divided broadly into four classes: class 1, neutralizing antibodies that block the ACE2 receptor and bind with the RBD in the closed state; class 2, neutralizing antibodies that block the ACE2 receptor and can bind to the RBD in both the open or closed states; class 3, neutralizing antibodies that bind outside the ACE2 site; and class 4, which bind with other sites and are generally less potent. Two, three, and two known monoclonal antibodies were selected from classes 1-3, respectively, each isolated from the sera of COVID-19 patients.
A yeast-display deep mutational scanning approach was utilized to generate a library of 3,800 SARS-CoV-2 RBD mutants, sorted to remove variants with no affinity for the ACE2 receptor as any such mutation eliminates the viability of the viral strain. Fluorescence-activated cell sorting was utilized to separate yeast bearing any of the antibodies employed from those without, allowing the mutations responsible for lessened affinity to be identified.
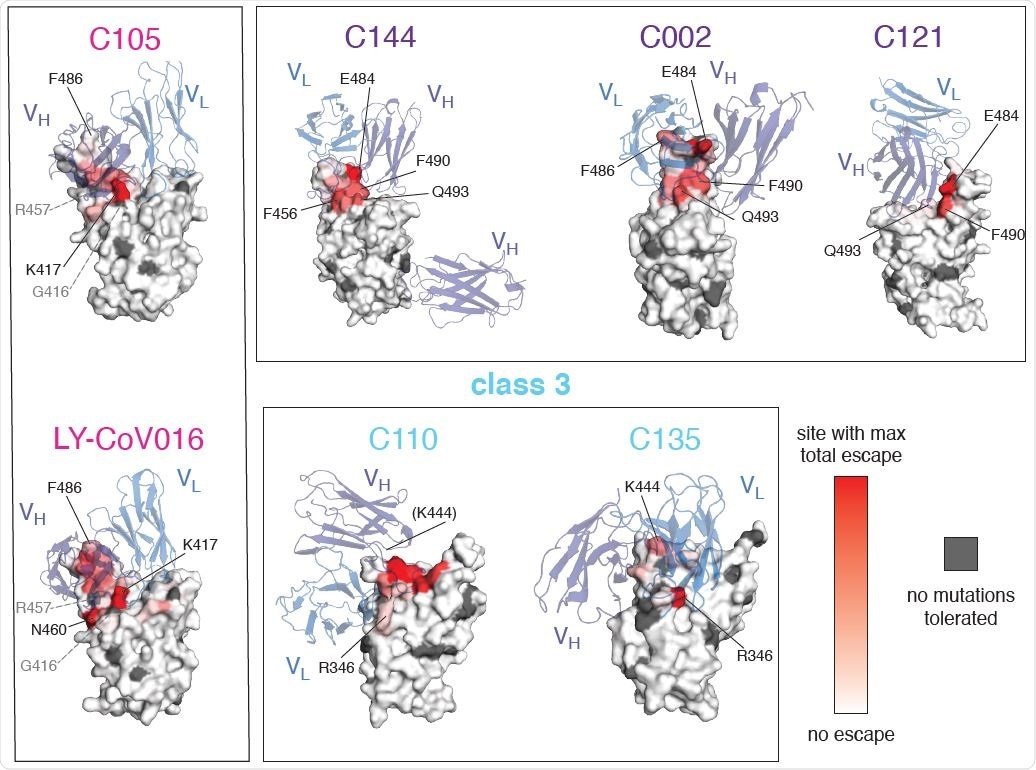
Mutations that escape antibody binding are usually in the direct structural footprint. (A) The total escape at each site is mapped onto the surface of the Fab-bound RBD, with white indicating no escape and red indicating the site with the most escape from that antibody. Sites where no mutations are tolerated for RBD folding or ACE2 binding are indicated in dark gray. For C105 and LY-CoV016, gray labels with dashed lines indicate example contact sites with no tolerated mutations. For C110, the general area where site 444 (unresolved in structure) would be located is indicated.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Which mutations allow escape?
The group found that mutations allowing antibody escape were generally found at similar sites in the RBD. Class 1 and 2 antibodies, in particular, were escaped by mutations in the RBD, though at distinct sites, while class 3 antibodies were escaped by a further non-RBD set of mutations. As both class 1 and 2 antibodies target regions of the RBD with some overlap, some mutations were observed to allow evasion from both.
Upon investigating the structure of the mutant RBDs that allowed antibody escape it was found that most were located at sites of direct contact with the antibody, directly providing steric hindrance that prevents successful conjugation. However, some sites of mutation not in direct contact with the antibody were also able to enhance evasion, either by perturbing the overall structure of the RBD or by charge-interactions that make close contact for bonding difficult.
The most prominent mutations for antibody evasion across the range of antibodies tested included those at sites F456 and E484, particularly notable as the class 2 antibody escaping E484K mutation has been documented in a number of highly prolific lineages, including B.1.351, P.1, P.2, and B.1.526. Of these, the B.1.351 and P.1 lineages also bear class 1 antibody escape mutation K417N or K417T, respectively. Very few lineages of note have been found to bear a class 3 antibody escape mutation, one example being the B.1.429 lineage, bearing the L452R mutation. As class 3 antibodies are generally less potent than class 1 or 2, this mutation has only a mild but still significant influence on immune escape. Currently, however, no strain incorporates mutations that allow escape from all three antibody classes.
Interestingly, some mutations had a profound influence against one or more of the monoclonal antibodies employed in this study, but little effect against the polyclonal antibodies, or vice versa. In any case, overall, mutations that allow escape from class 2 antibodies were found to be the most effective. As many class 1 antibodies share binding sites with class 2 antibodies, these mutations were effective against both, and class 2 antibodies also contributed most strongly to evasion from polyclonal plasma. Indeed, the previously documented class 2 antibody escape mutation E484K was demonstrated to lessen polyclonal plasma neutralization by around five times.
The group suggests that the ability of class 2 antibodies to target the RBD both in the open or closed conformations explains the dominance of this class in the immune response, though that it is this specificity that drives the rapid evolution of SARS-CoV-2 lineages able to escape these antibodies. Therefore, close and careful monitoring of the development of such mutations is of vital importance to ongoing public health and vaccine development efforts.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mutational escape from the polyclonal antibody response to SARS-CoV-2 infection is largely shaped by a single class of antibodies, Allison J. Greaney, Tyler N. Starr, Christopher O. Barnes, Yiska Weisblum, Fabian Schmidt, Marina Caskey, Christian Gaebler, Alice Cho, Marianna Agudelo, Shlomo Finkin, Zijun Wang, Daniel Poston, Frauke Muecksch, Theodora Hatziioannou, Paul D. Bieniasz, Davide F. Robbiani, Michel C. Nussenzweig, Pamela J. Bjorkman, Jesse D. Bloom, bioRxiv, 2021.03.17.435863; doi: https://doi.org/10.1101/2021.03.17.435863, https://www.biorxiv.org/content/10.1101/2021.03.17.435863v1
- Peer reviewed and published scientific report.
Greaney, Allison J., Tyler N. Starr, Christopher O. Barnes, Yiska Weisblum, Fabian Schmidt, Marina Caskey, Christian Gaebler, et al. 2021. “Mapping Mutations to the SARS-CoV-2 RBD That Escape Binding by Different Classes of Antibodies.” Nature Communications 12 (1). https://doi.org/10.1038/s41467-021-24435-8. https://www.nature.com/articles/s41467-021-24435-8.