The COVID-19 pandemic remains a major global crisis that has led to extensive adverse health and socioeconomic impacts. Although SARS-CoV-1, SARS-CoV-2, and MERS-CoV viruses belong to the betacoronavirus genus, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seems to have a greater transmission rate compared to previous outbreaks of betacoronaviruses. SARS-CoV-2 spread rapidly to almost all countries worldwide, with continued transmission promoted by new variants of the virus.
Given the emergence of the UK, South African, and South American variants of SARS-CoV-2 and the likelihood of further mutations, there is a compelling need to develop efficient strategies to prevent and manage such outbreaks. A thorough understanding of the mechanism of SARS-CoV-2 infection starting with the virus-host cell interaction is crucial to develop these prevention strategies. Previous studies have shown that several molecular-level factors contribute to increased SARS-CoV-2 infectivity compared to that of SARS-CoV-1.
Based on high-resolution structures, these coronaviruses use a similar mechanism for entry into the host cell, and the initial infection is very much dependent on host cell recognition. This is mediated by the viral S (spike) protein with the help of the human angiotensin-converting enzyme 2 (ACE2). While the ACE2 is a cell surface protein present in various tissues, the spike protein is a homotrimeric type I fusion glycoprotein that protrudes from the viral surface and has two subunits, S1 and S2. Since this spike protein-ACE2 (protein-protein) interaction helps initiate infection, the viral spike protein is a very important therapeutic target.
Comparing the relative dynamics between the SARS-CoV RBD-ACE2 and SARS-CoV-2 RBD-ACE2 complexes
Recently, researchers from the US employed multiple-µs molecular dynamic simulations to compare the relative dynamics, stabilities, and energy differences between the SARS-CoV receptor-binding domain (RBD)-ACE2 and SARS-CoV-2 RBD-ACE2 complexes. The study is published on the preprint server, bioRxiv*.
Recent advances in hardware and software allow molecular dynamics simulations on increasingly larger systems over longer timescales than were previously possible. These simulations produce large amounts of data, posing critical challenges for data analysis and interpretation. To tackle this, machine learning approaches were used to help identify residues that contribute the most to dynamical differences in the two viral RBDs.
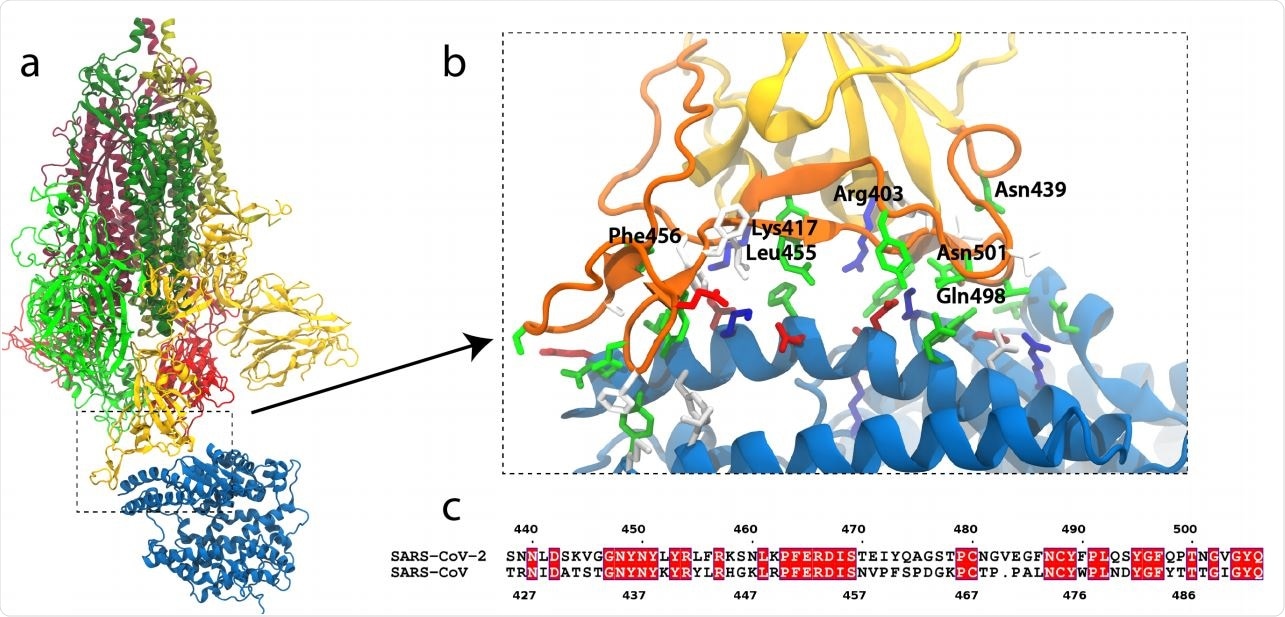
SARS-CoV-2 S-protein binding to ACE2. (a) S-protein trimer (green, yellow, red) bound to the peptidase domain of ACE2 (blue). The lighter and darker shades of the trimer are S1 and S2, respectively. The viral envelope would be at the top and the host cell membrane at the bottom. (b) Close-up of the RBM (orange) interacting with ACE2. Residues near the interface are shown in a stick representation, colored by residue type (blue and red are positively and negatively charged, respectively, green is polar, white is hydrophobic). Specific residues in SARS-CoV-2 are labeled. (c) Alignment of the RBMs of SARS-CoV-2 and SARS-CoV19. Identical residues are white on red background.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Machine learning approaches accurately identified the most meaningful mutations for subsequent FEP calculations
The researchers used molecular dynamic simulations, machine learning, and free energy perturbation (FEP) calculations to illustrate differences in RBD binding by the two viruses. Although they observed only subtle differences from the initial molecular dynamic simulations of the 2 RBD-ACE2 complexes, machine learning identified the individual residues with distinctive ACE2 interactions, many of which were already highlighted by previous studies.
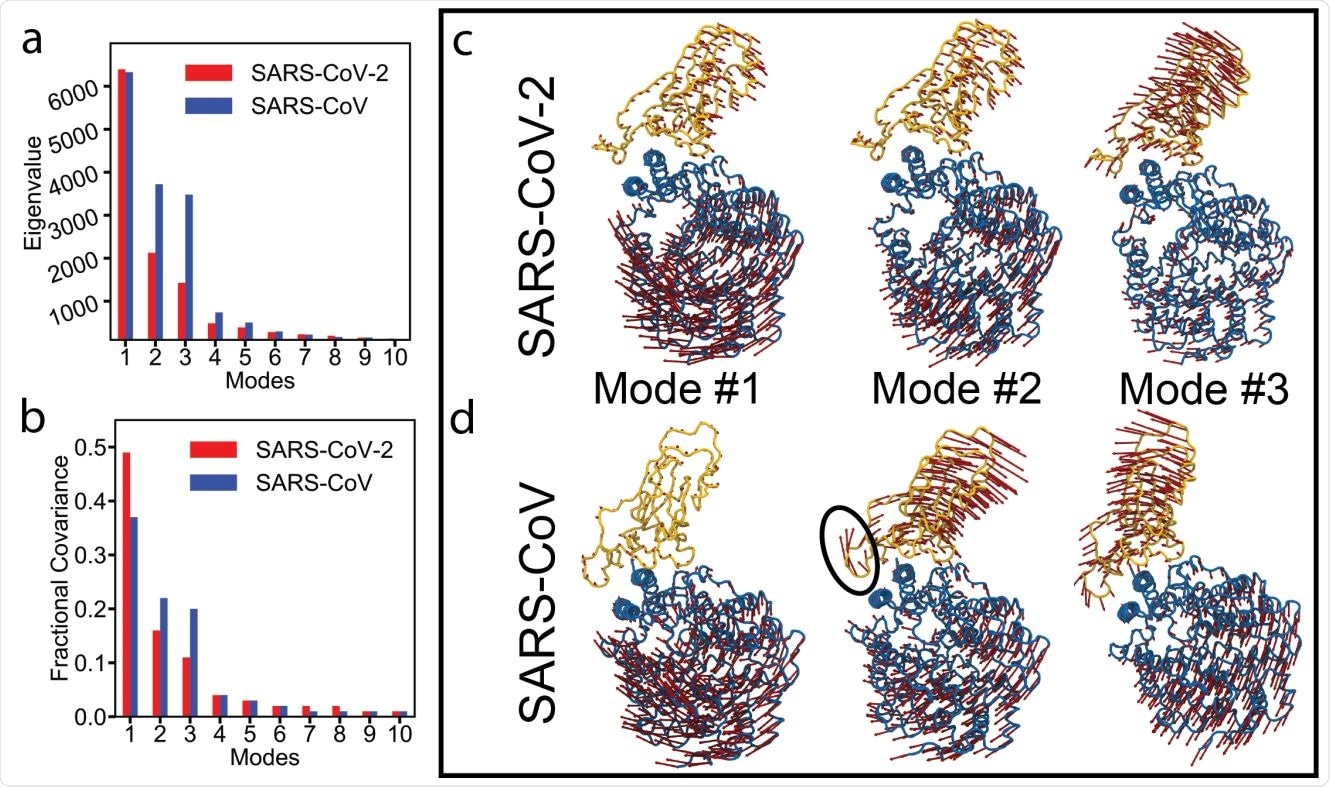
Essential dynamics analysis of two RBD-ACE2 complexes of SARS-CoV-2 and SARS-CoV viruses. (a) Eigenvalues and (b) fractional variance of top-10 modes. Three dominant modes of the complexes with (c) SARS-CoV-2 and (d) SARS-CoV. ACE2 and RBD are shown in blue and yellow, respectively. Motion of a loop in SARS-CoV in mode #2 is highlighted with a black oval.
FEP calculations measured the corresponding differences in binding free energies to ACE2, and analysis of MD trajectories offered structural explanations for these observed differences. The energetics of emerging SARS-CoV-2 mutations were also studied, and it showed that the RBD’s affinity for ACE2 is increased by the N501Y and E484K mutations and slightly decreased by the K417N mutation.
“Significant differences in free energies (>1 kcal/mol) were observed for five of the seven mutations selected by our ML approaches, indicating that they accurately identified the most meaningful mutations for subsequent FEP calculations.”
The study demonstrates the utility of machine learning approaches in analyzing long MD trajectories
According to the authors, the study results suggest that the effects on ACE2 binding play a significant role in the natural selection of mutations in the RBD of SARS-CoV-2. Also, the mutations that affect ACE2 binding are not likely to become widespread. This work demonstrates the utility of machine learning approaches in analyzing long MD trajectories that could be readily generalized to other complexes.
“Our FEP results are in agreement with deep mutational scanning experiments by Star et al. as well as other experimental assays, demonstrating that our approach is a fast, accurate way to predict the effect of emerging SARS-CoV-2 mutations in the RBD.”
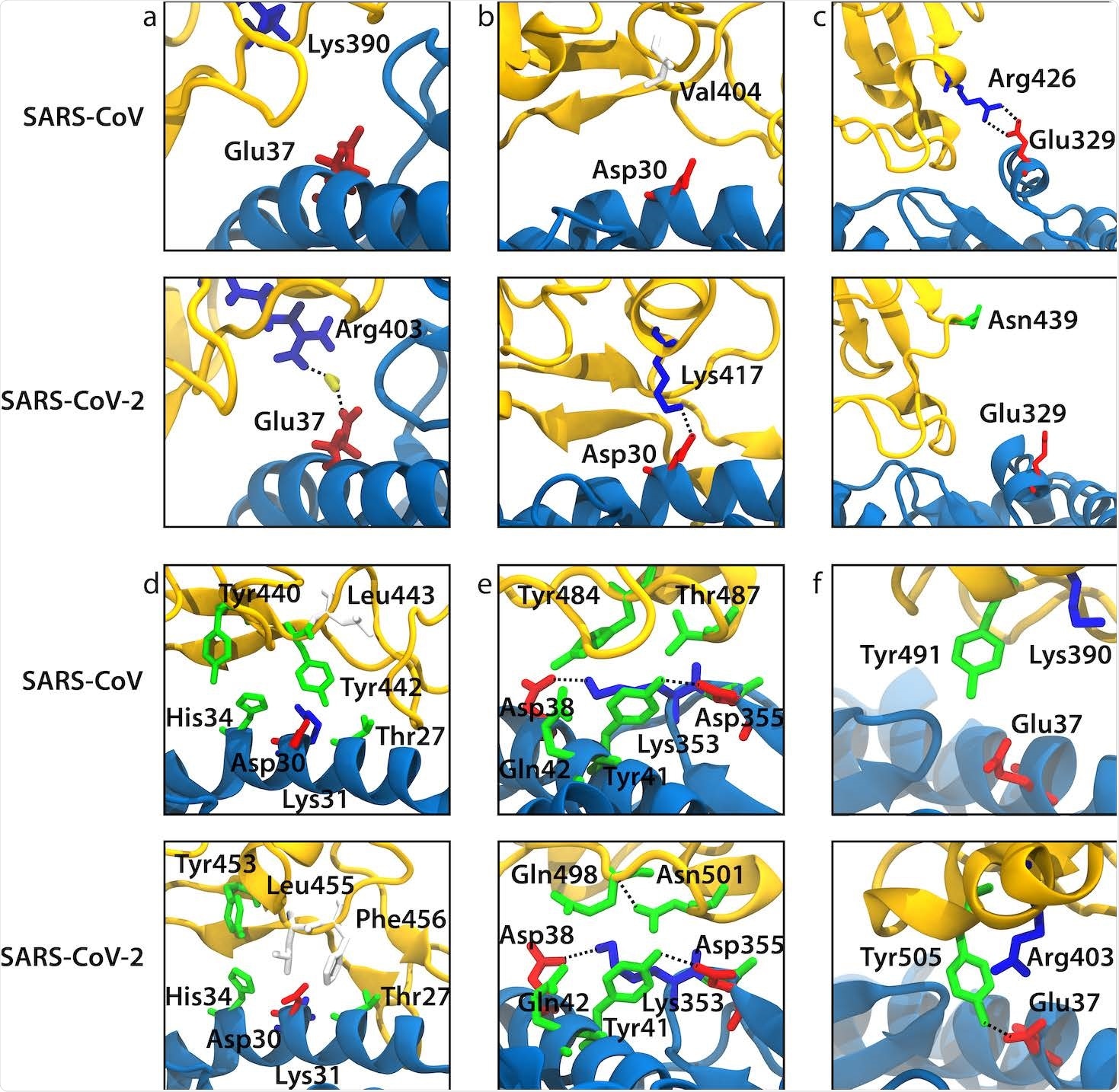
Snapshots from our MD simulations that illustrate the difference in ACE2 binding between SARS-CoV and SARS-CoV-2 for the selected mutations. ACE2 and RBD backbones are shown in blue and yellow, respectively. (a) Arg403/Lys390 (SARS-CoV-2/SARS-CoV numbering). (b) Lys417/Val404. (c) Asn439/Arg426. (d) Leu455/Tyr442. (e) Gln498-Asn501/Tyr484-Thr487. (f) Tyr491/Tyr505.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Critical interactions for SARS-CoV-2 spike protein binding to ACE2 identified by machine learning, Anna Pavlova, Zijian Zhang, Atanu Acharya, Diane L Lynch, Yui Tik Pang, Zhongyu Mou, Jerry M Parks, Chris Chipot, James C. Gumbart, bioRxiv, 2021.03.19.436231; doi: https://doi.org/10.1101/2021.03.19.436231, https://www.biorxiv.org/content/10.1101/2021.03.19.436231v1
- Peer reviewed and published scientific report.
Pavlova, Anna, Zijian Zhang, Atanu Acharya, Diane L. Lynch, Yui Tik Pang, Zhongyu Mou, Jerry M. Parks, Chris Chipot, and James C. Gumbart. 2021. “Machine Learning Reveals the Critical Interactions for SARS-CoV-2 Spike Protein Binding to ACE2.” The Journal of Physical Chemistry Letters 12 (23): 5494–5502. https://doi.org/10.1021/acs.jpclett.1c01494. https://pubs.acs.org/doi/10.1021/acs.jpclett.1c01494.