Previous research identified the interferon-responsive OAS1 gene as a risk gene for Alzheimer's disease. OAS1 is an enzyme that binds to double-stranded RNA and induces the conformation to create oligoadenylates. They also activate RNase L to break down RNA.
Researchers led by Dervis A. Salih from the UK Dementia Research Institute at UCL have recently expanded on these findings. They found that decreases in OASI gene expression were associated with the development of Alzheimer's disease. Low expression from an OAS1 variant also promoted an inflammatory response, which could contribute to severe symptoms in COVID-19 illness.
The authors write:
"Our data show that OAS1 is required to limit the pro-inflammatory response of myeloid cells when stimulated with IFN-γ. We also identify a SNP within OAS1 associated with AD in the same locus that predisposes to critical illness with COVID-19…Further investigation of the function of OAS1 in innate immune cells and the genetic network engaged by OAS1 will provide better molecular targets to track disease progression and treat AD, as well as COVID-19 and potentially its long-term sequelae."
The results could aid with future therapeutics for Alzheimer's and COVID-19 by treating OASI variants. The risk gene could also be beneficial in predicting the onset of disease.
The study "Genetic variability associated with OAS1 expression in myeloid cells increases the risk of Alzheimer's disease and severe COVID-19 outcomes" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
rs1131454 within OAS1 is linked to Alzheimer's and severe COVID-19 infection
The research team genotyped 1,313 participants with sporadic Alzheimer's disease and 1,234 participants who served as controls. Genotyping results identified rs1131454 in the OAS1 gene is linked to Alzheimer's disease. The presence of rs1131454 was associated with severe COVID-19 illness, indicating the gene variant affects both Alzheimer's disease and SARS-CoV-2 infection.
The team then used scRNA-seq of isolated mouse microglia to build genetic transcriptome networks. They uncovered an interferon response pathway with increased expression from Oas1a during aging in microglia. The findings suggest the interferon pathway happens in a subpopulation of innate immune cells. Changes in genetic expression from aging could thus increase or decrease the risk of developing age-related diseases.
OAS1 variant influences the inflammatory response
Autopsies of individuals who died from Alzheimer's showed interferon-responsive genes with OAS1 in human microglia. Using human microglia-like genes, they showed the OAS1 gene was the culprit behind the pro-inflammatory response elicited by myeloid cells when exposed to increased interferon levels.
Decreased OAS1 levels were more likely to exhibit a strong pro-inflammatory response and were linked to Alzheimer's and COVID-19. The researchers suggest the eQTL variants contributed to lower OAS1 levels. Individuals with the eQTL variant on the OAS1 gene have an increased likelihood of damaging the neurons and alveolar cells by triggering a cytokine storm.
Using siRNA, the team lowered OAS1 expression to mimic the eQTL variant. They found the gene does play an essential role in mitigating the pro-inflammatory marker TNF-α when the levels of IFN-γ increase.
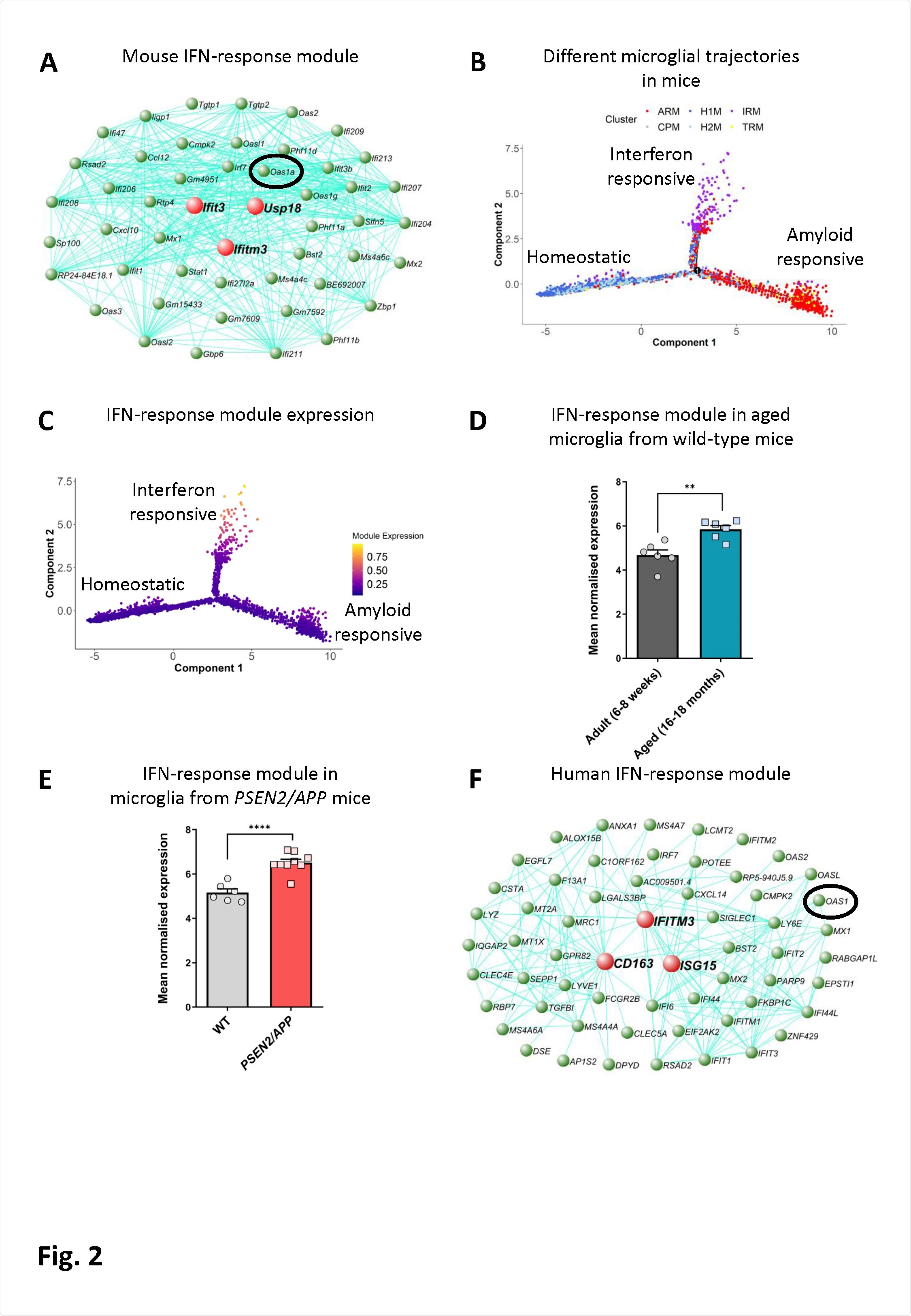
An interferon-response-associated gene module is present along a distinct microglial activation trajectory upregulated in aged mice, mice with amyloid pathology and in humans with AD. A) The genetic network containing Oas1a from microglial cells isolated from wild-type and APPNL-G-F KI mice at 3, 6, 12 and 21 months of age analyzed by scRNA-seq (Sala Frigerio et al., 2019). The 50 genes showing the highest connectivity are plotted, and Oas1a is highlighted. Green nodes represent genes, edge lines represent coexpression connections, and the central large red nodes are the hub genes (full network given in Table S2). (B) Semi-supervised pseudotime ordering of microglial cells isolated from wild-type and APPNL-G-F KI mice as above based on expression (Sala Frigerio et al., 2019), with Monocle 2, shows homeostatic cells as the root state, and ARM and IRM as the endpoints of distinct activation trajectories. C) The gene module containing Oas1a is upregulated along the IRM-associated activation trajectory. The expression of this module is relatively absent from both the root homeostatic state and the ARM trajectory. D) Mean normalized expression of the 60 most central genes in the interferon response module is greater in microglia isolated from aged wild-type relative to young adult mice (6-8-weeks versus 16-18-months of age; N=6 mice per group. Data are shown as mean ± SEM. Student’s test; **p<0.01. Further analysis of data from O’Neil et al. (2018)). E) Mean normalized expression of the 60 most central genes in the interferon response module is greater in microglia isolated from PSEN2/APP relative to wild-type mice at 14-15-months of age (N=6- 9 mice per group. Data are shown as mean ± SEM. Student’s t-test; ****p<0.0001. Further analysis of data from Friedman et al. (2018)). F) Genetic network plot of an interferon response associated module detected in microglial cells isolated from human AD patients and individuals with MCI (Olah et al., 2020). This module shows a significant overlap with the interferon-response module detected in mice (see panel A) (full network given in Table S3). The hub gene of this module is CD163, and a number of other macrophage marker genes are prominent within this module (MSR1, TGFB1, F13A1, LY6E and LYZ), indicating that expression of OAS1 and other interferon-response genes in the human AD patient brain are either associated with a population of CNS border-associated or invading macrophages, or potentially a microglial subpopulation which upregulates anti-inflammatory genes. OAS1 is highlighted.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Increased inflammation suppresses TREM2 expression
The team also observed that elevated levels of IFN-γ and TNF-α prevented the expression of TREM2.
"Given that TREM2 is likely to have a protective role in AD and slow disease progression, elevated pro-inflammatory signals could increase the risk and progression of AD by suppressing this protective signal. TNF-α also activates PKR/ EIF2αK2, which is downstream of interferon signaling via TLR4, and consequently the protein activator of the IFN-inducible protein kinase (PRKRA) leading to apoptosis," wrote the researchers.
Their findings suggest high levels of IFN signaling coupled with TNF-α release could be the mechanism behind the development of dementia.
Although, upregulation of interferon-responsive genes — Oas1a and family members Mx1, Stat1/2, Ifit3, Ifitm3, and Usp18 — with ages could limit the amount of damage induced by age-related pro-inflammatory signaling. Considering old age is a high-risk factor for severe COVID-19 infection and death, the researchers suggest an understanding of changes with OAS1a expression could help mitigate the age-related cellular damage that contributes to diseases' development.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Magusali N, et al. Genetic variability associated with OAS1 expression in myeloid cells increases the risk of Alzheimer's disease and severe COVID-19 outcomes. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.16.435702, https://www.biorxiv.org/content/10.1101/2021.03.16.435702v1
- Peer reviewed and published scientific report.
Magusali, Naciye, Andrew C Graham, Thomas M Piers, Pantila Panichnantakul, Umran Yaman, Maryam Shoai, et al. 2021. "A genetic link between risk for Alzheimer's disease and severe COVID-19 outcomes via the OAS1 gene." Brain, 144(12). https://doi.org/10.1093/brain/awab337. https://academic.oup.com/brain/article/144/12/3727/6382473.