The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic and the vast societal and economic consequences of the connected lockdowns have had devastating consequences for the world's populations. Despite rapid vaccine development progress, some emerging virus variants appear to be less effectively neutralized by antibodies elicited by the first-generation vaccines, and the emergence of further variants is expected. To date, over 124 million people have been infected with this virus globally.
As the scientific community continues its search for new SARS-CoV-2 vaccines and therapeutic agents for the clinical management of the COVID-19 disease, several new variants of the original virus have emerged that appear to cause significant changes in the way the pathogen acts, including alterations to its contagiousness. Some of the variants are 20I/501Y.V1, VOC 202012/01, or B.1.1.7 (in the UK), 20H/501Y.V2 or B.1.351 (in South Africa), and P.1 (in Brazil and also in the US).
Scientists suspect that the variants of the SARS-CoV-2 result in changes in its transmissibility, clinical presentation, and severity, or impact countermeasures, including diagnostics, therapeutics, and vaccines. Studies reported that some emerging virus variants appear less effectively neutralized by antibodies elicited by the first-generation vaccines. To arrest virus escape (because of these emerging variants), researchers are exploring distinct and novel neutralization mechanisms.
In a new study, published recently on the bioRxiv* preprint server, researchers isolated a nanobody that potently neutralizes SARS-CoV-2, including the B.1.351 variant, and cross-neutralizes SARS-CoV. They demonstrated the therapeutic potential of the nanobody in a human ACE2 (angiotensin-converting enzyme) transgenic mouse model.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Using biochemistry and electron cryomicroscopy, they showed that this nanobody simultaneously interacts with the two RBDs (receptor binding domains) from different spike trimers, rapidly inducing the formation of spike trimer-dimers. This naturally elicited bispecific monomeric nanobody establishes a novel strategy for potent immobilization of viral antigens
The viral spike (S) protein enables the SARS-CoV-2 attachment to the human host cell via the ACE2 receptor. In the prefusion conformation, the spike is a large homotrimer; each S monomer consists of two subunits: S1 and S2. The S1 subunit contains the RBD and the S2 contains the fusion peptide, as well as the transmembrane domain.
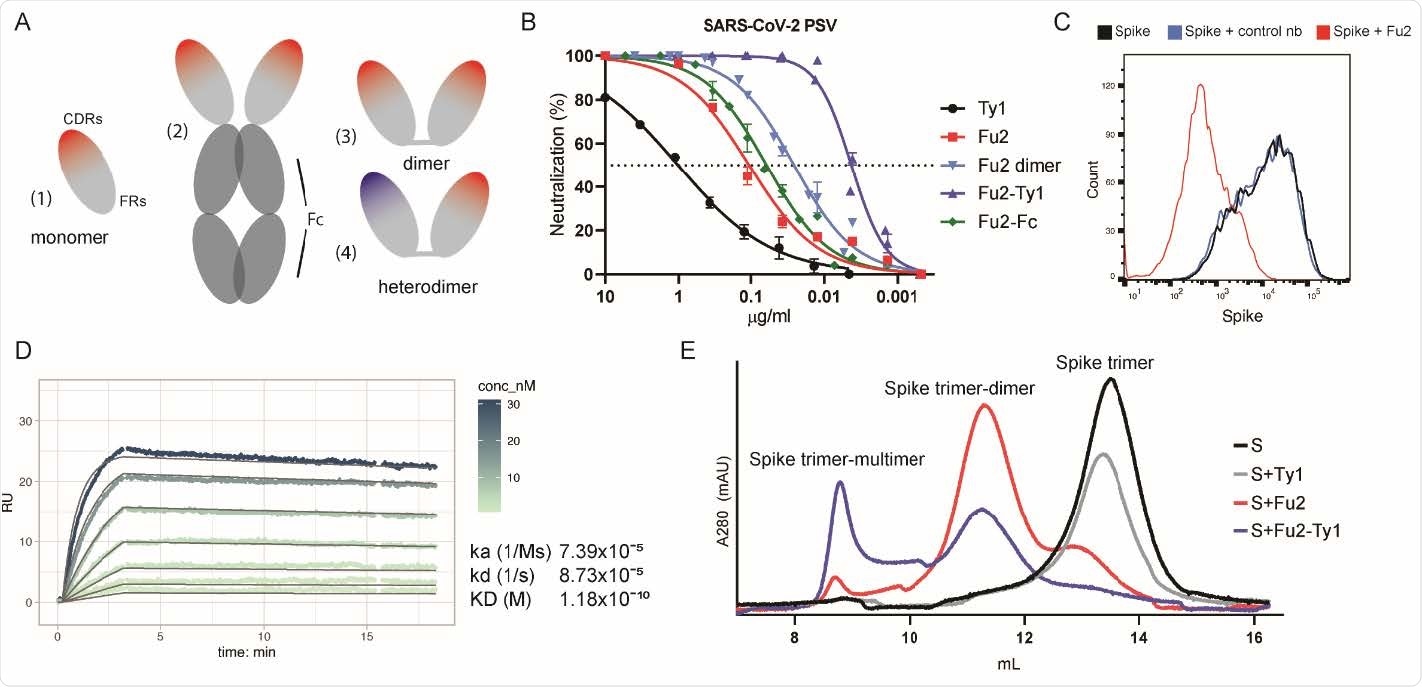
An RBD-specific nanobody neutralizes SARS-CoV-2. (A) Overview of different nanobody constructs used in this study: (1) nanobody monomer, (2) nanobody Fc-fusion, (3) chemically linked nanobody homodimer, (4) chemically linked nanobody heterodimer. (B) A SARS-CoV-2 spike pseudotyped lentivirus (PSV) was incubated with a dilution series of the indicated nanobodies at 37°C for one hour before infecting HEK293T-hACE2 cells. Neutralization (in %) compared to untreated PSV is shown. Data from at least two replicates is shown and the error bars represent the standard deviation. (C) Recombinantly expressed, prefusion stabilized, and fluorescently labeled SARS-CoV-2 spike protein was incubated with a control nanobody specific for IAV NP 29 274 , or Fu2 and used to stain ACE2 expressing HEK293T cells. Cells were analyzed by flow cytometry and a representative histogram is shown. (D) Binding kinetics of Fu2 to the RBD were measured by surface plasmon resonance (SPR). Sensorgram is color-coded based on concentration. The fit is based on the 1:1 Langmuir model and is shown in dark grey solid lines. (E) Recombinantly expressed, prefusion stabilized SARS279 CoV-2 spike protein was run alone or preincubated with the indicated nanobody constructs on a Superose 6 size-exclusion column. Elution profiles (A280) are shown.
The SARS-CoV-2 is quickly neutralized by high-affinity binding antibodies and other target molecules that bind to, or close to, the receptor-binding motif of the RBD and neutralize by directly hindering virus attachment.
Camelid-derived single-domain antibody fragments - called the nanobodies - are promising candidates as therapeutic agents in the early stages of SARS-CoV-2 infection. The nanobodies have high specificity, affinity, and stability; it is easy and affordable to produce in large scales. Nanobodies have high potency - can target multiple epitopes simultaneously, by forming homo- and heterodimers or other multimers. Thus, the use of nanobodies allows the neutralization of multiple known or unknown virus variants and reduces the risk of viral escape.
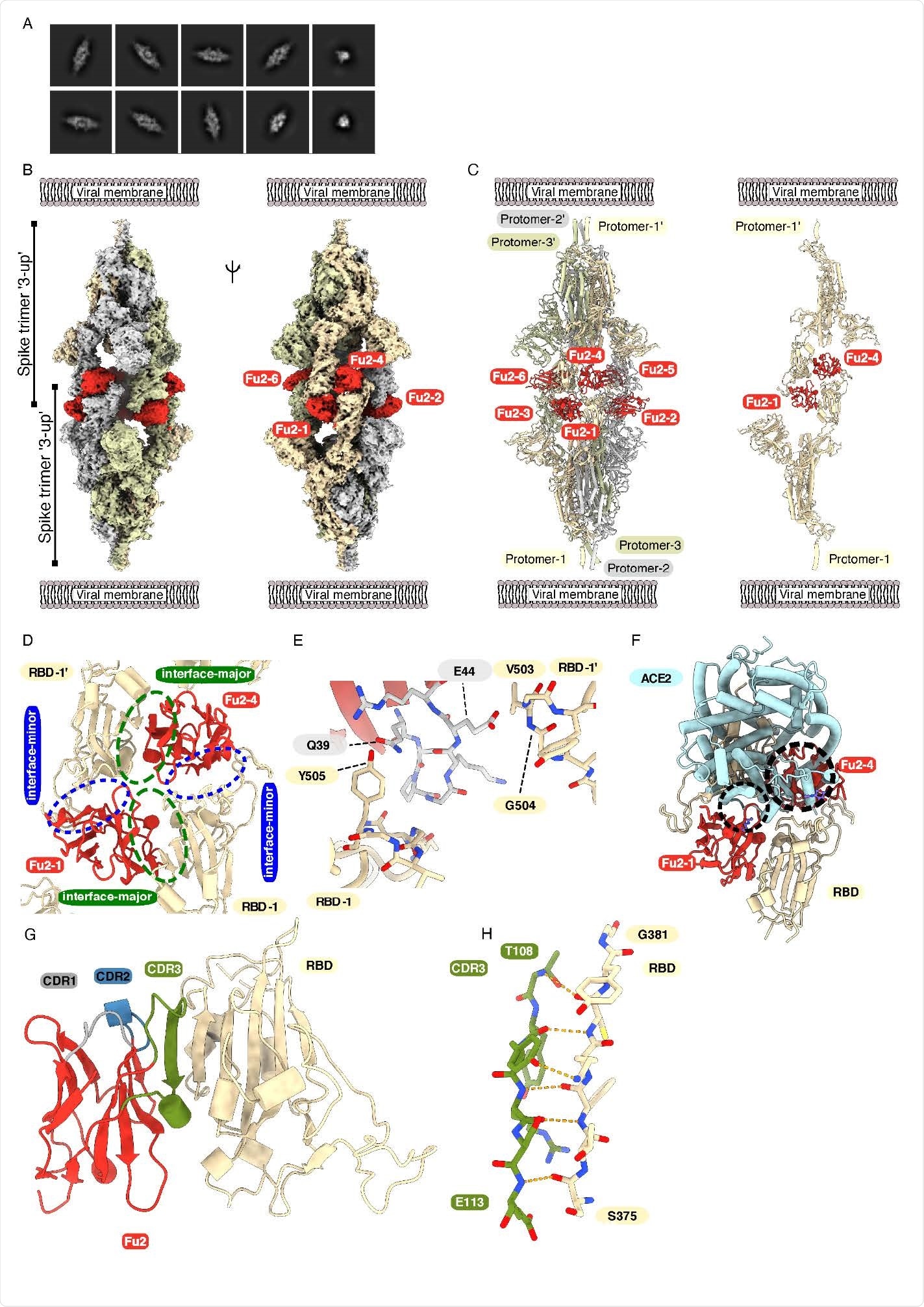
Cryo-EM provides structural evidence of Fu2-mediated dimerization of trimeric spike in ‘3-up’ conformation (A) Representative 2D class averages. (B) Cryo-EM map (two different side views) and (C) Atomic model of ‘dimeric’ spike in complex with Fu2; and two opposite protomers with two Fu2 molecules. Both spike trimers in the complex are present in ‘3- up’ conformation (six Fu2 molecules are numbered). (D) Close-up view of the RBD-Fu2 interaction interfaces showing the interface-major and interface-minor. (E) A b-hairpin (gray sticks) from the Fu2 framework region (residues 39-45) contacts two RBDs simultaneously in the Fu2-mediated spike trimer-dimer. (F) Modeling of the simultaneous spike-Fu2-ACE2 binding shows that Fu2 blocks the binding of ACE2 to the RBD from two directions. Clashes marked with dashed circles. (G) Fu2 binds the RBD through an extended inter-chain b-sheet interaction mediated by the CDR3 region (green). CDR1 (gray) and CDR2 (blue) does not take part in RBD binding (H) Closeup of a part of the CDR3-RBD interface displaying the anti-parallel b-strand hydrogen bond pairing between the Fu2 CDR3 region and the RBD (light orange).
Because of the emergence of new virus variants that evade antibody-mediated neutralization, it is crucial to identify molecules such as nanobodies with a more diverse set of binding epitopes and neutralization mechanisms that allow for the neutralization of a variety of virus variants. In this study, the researchers have generated a nanobody with two non-overlapping interaction sites on the SARS-CoV-2 RBD, each targeting a different epitope, which induces the formation of dimers of spike trimers and eventually suppresses the SARS-CoV-2 in vivo.
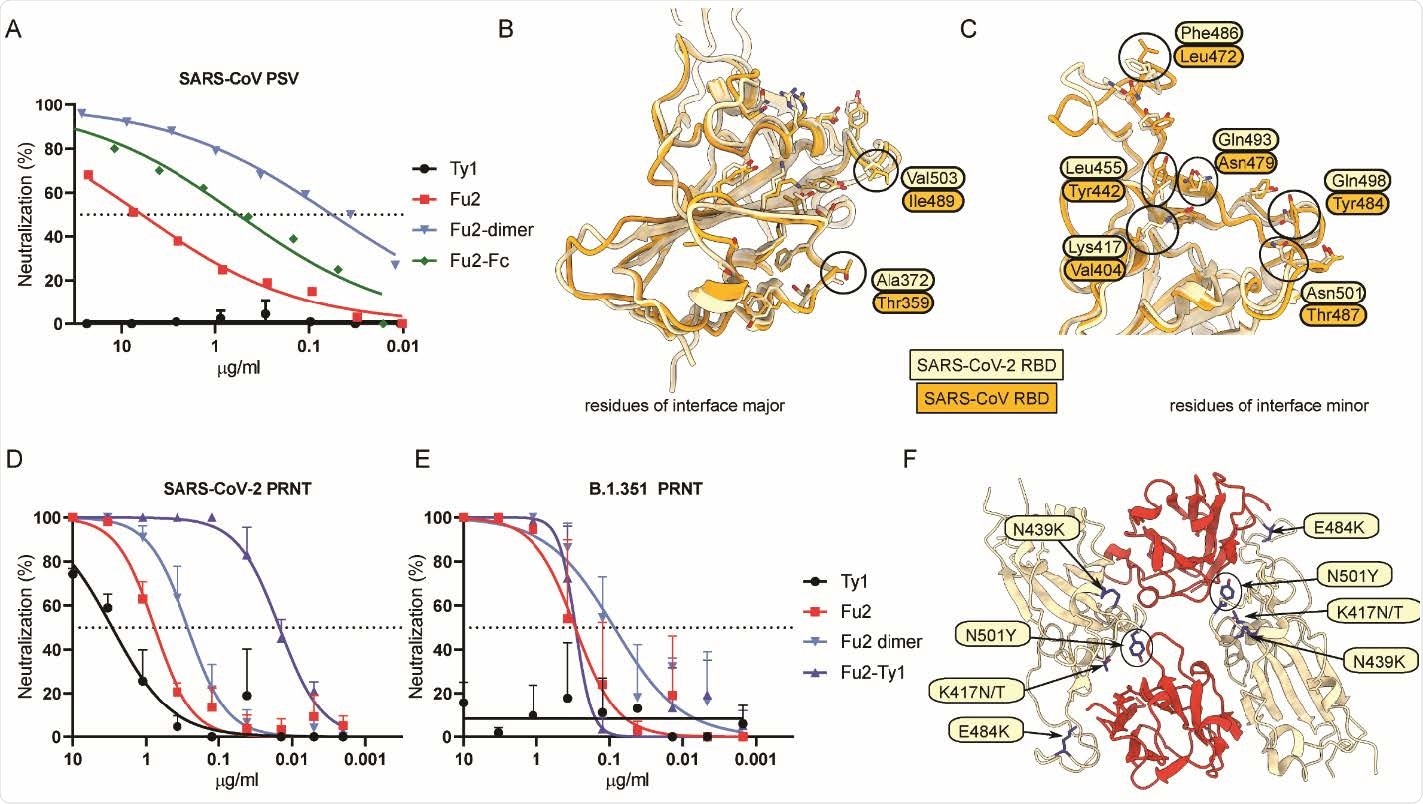
Fu2 cross-neutralizes SARS-CoV and a B.1.351 variant of concern clinical isolate (A) SARS-CoV S pseudotyped lentivirus (PSV) was incubated with the indicated constructs before infecting HEK293T-ACE2 cells, and neutralization is shown compared to untreated PSV. (B and C) Structural basis of SARS-CoV cross-neutralization by Fu2. Structural alignment of SARS-CoV-2 Spike (RBD bound Fu2) with SARS-CoV RBD (PDB:6ACD). Residues of interface-major and interface-minor shown as sticks, divergent residues are labeled. (D and E) PRNTs with clinical isolates of SARS-CoV-2. 100 plaque forming units (PFU) of SARS-CoV-2 18 or B.1.351 (501Y.V2) 1 were incubated with dilution series of the indicated nanobody constructs for 1 h at 37°C before infecting monolayers of Vero E6 cells. Plaques were quantified 72 h post infection. Neutralization representing the reduction of plaques relative to the control wells is shown. Error bars represent standard deviation (SD) across replicate experiments. (F) Assessment of Fu2-RBD interaction in RBD variants. In silico mutational analyses of RBD residues and probable impact on Fu2 binding. Fu2 is shown in red and variant residues are indicated.
The researchers described that the novel nanobody in this study, Fu2, can effectively neutralize the B.1.351 variant. Importantly, they showed that Fu2 can neutralize the SARS-CoV, highlighting its specificity for a more conserved epitope. Compared to the monomeric nanobody, the dimeric constructs had a better affinity to the RBD, presenting significantly increased potency.
The researchers also reported a dose-dependent study of the nanobody (serum half-lives ranges ~1-3 hours) in the hACE2 transgenic mice. Instead of a high dose of the nanobody (which did not reduce the COVID-19 severity), the heterodimer was administered daily for four days post-challenge. They observed a significant delay in the disease onset and severity, suggesting that nanobodies and dimeric nanobody constructs have the potential for SARS-CoV-2 antiviral therapy.
The interaction between Fu2 and the RBD is highly unusual - two copies of an antigen must be bound at different epitopes with precisely the right spatial arrangement.
"While this novel mode of antigen-binding may be uncommon in naturally elicited antibody repertoires, it could be achievable by structure-based antibody design. It might present a general strategy for potent immobilization of viral glycoproteins and other antigens."
The present scenario and possible future emergencies, alternatives and novel ways to inhibit and neutralize the virus and variants of concern are needed. This study has demonstrated that the nanobody has a unique and robust potential to form the basis for developing therapeutic interventions against this emerging SARS-CoV-2 variant of concern.
It remains to be seen whether 'designer' bispecific monomer nanobodies can push the potency envelope beyond what is currently attainable, the researchers signed off.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A bispecific monomeric nanobody induces SARS-COV-2 spike trimer dimers Leo Hanke, Hrishikesh Das, Daniel J Sheward, Laura Perez Vidakovics, Egon Urgard, Ainhoa Moliner-Morro, Vivien Karl, Alec Pankow, Changil Kim, Natalie L Smith, Gabriel K Pedersen, Jonathan M Coquet, B Martin Hällberg, Ben Murrell, Gerald M McInerney bioRxiv 2021.03.20.436243; doi: https://doi.org/10.1101/2021.03.20.436243, https://www.biorxiv.org/content/10.1101/2021.03.20.436243v1
- Peer reviewed and published scientific report.
Hanke, Leo, Hrishikesh Das, Daniel J. Sheward, Laura Perez Vidakovics, Egon Urgard, Ainhoa Moliner-Morro, Changil Kim, et al. 2022. “A Bispecific Monomeric Nanobody Induces Spike Trimer Dimers and Neutralizes SARS-CoV-2 in Vivo.” Nature Communications 13 (1): 155. https://doi.org/10.1038/s41467-021-27610-z. https://www.nature.com/articles/s41467-021-27610-z.