A new fast-spreading severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant called B.1.526 recently emerged in New York City and caused concerns for its potential to resist the immune system.
Takuya Tada from NYU Grossman School of Medicine led a research team to test how susceptible the B.1.526 variants were to neutralizing antibodies from convalescent plasma, monoclonal antibodies, and mRNA vaccines.
They found the current coronavirus treatments such as therapeutic monoclonal antibodies and vaccine-induced neutralizing antibodies help protect against the B.1.526 variant with the S477N mutation. The treatment's neutralizing ability somewhat decreased against the B.1.526 variant with the E484K mutation.
"Our findings should assuage concerns that the B.1.526 variant will evade protection provided by vaccine-elicited antibodies and suggest that therapeutic antibody therapy will retain its effectiveness against the variant." However, the researchers suggest their results are further evidence for a need for wide-spread vaccination.
The study "B.1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Features of B.1.526 Variant
The B.1.526 was first detected in November 2020, but by January 2021, it comprised 5% of genomes sequenced in New York City neighborhoods and increased to 12.3% by mid-February.
Several versions of B.1.526 have emerged with several mutations on the spike protein's receptor-binding domain. One version of the B.1.526 variant has an E484K mutation — the same shared by B.1.351 and P1, which the World Health Organization has categorized as variants of concern. Another version lacks E484K but has an S477N mutation on the spike protein, which increases affinity for the ACE2 receptor.
Both versions have both the D614G mutation and A701V and L5F, T95I and D253G — mutations not observed in previously reported variants.
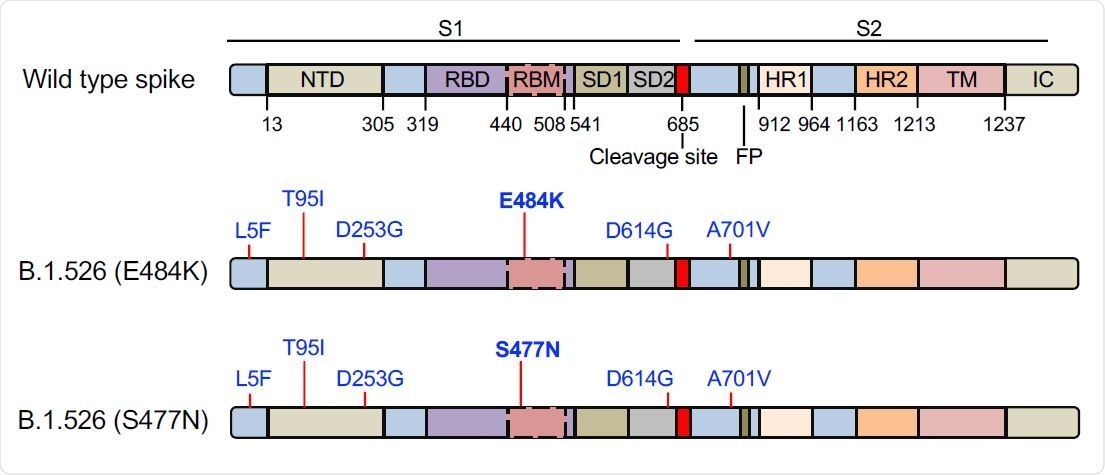
The domain structure of the SARS-CoV-2 spike protein is shown above. NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; SD1 subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane region; IC, intracellular domain. The location of the mutations in the two B.1.526 spike proteins is diagrammed below with the distinguishing E484K and S477N mutations in bold.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Convalescent sera versus B.1.526
The researchers constructed spike protein expression vectors for both B.1.526 versions to create lentiviral pseudotypes reporter viruses to study the B.1.526 spike proteins. Using immunoblot analysis, they found B.1.526 spike proteins were in transfected 293T cells. Both were at virion levels similar to the D614G spike protein most common in the original strain found in Wuhan.
The next step was to test how effective convalescent sera would be in neutralizing both B.1.526 variants. The convalescent sera were collected from individuals infected with SARS-CoV-2 before April 2020 and had either B.1.526, D614G, and E484K mutations on the coronavirus spike proteins.
Results showed the B.1.526 variant with the S477N mutation had neutralizing titers similar to SARS-CoV-2 with the D614G mutation. With the B.1.526 variant with an E484K mutation, the convalescent sera produced a modest decrease by 3.8-fold.
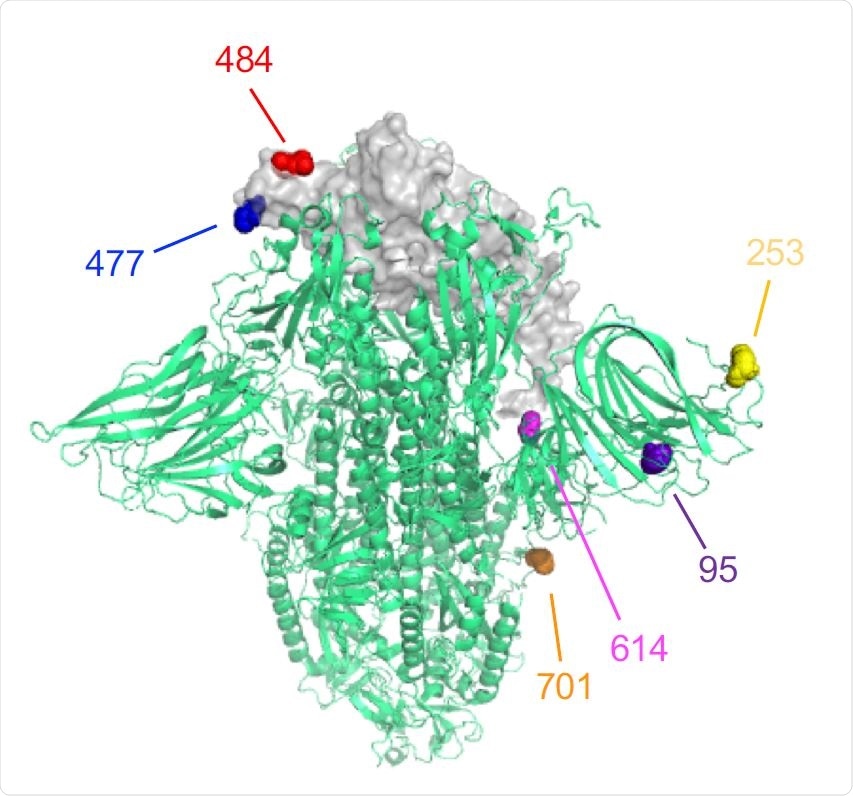
The location of the B.1.526 variant spike protein mutations is shown on the 3D structure of the trimeric spike protein. One RBD region is shown for simplicity. The 484 (red) and 477 (blue) amino acid residues are indicated.
Vaccine-induced antibodies versus B.1.526
The research team gathered neutralizing titers of serum from vaccinated individuals with either the Pfizer-BioNTech mRNA vaccine or the Moderna mRNA vaccine. They found the Pfizer vaccine produced antibodies that effectively neutralized D614G and the B.1.1.7 variant. However, there was a 3.4-fold decrease in neutralizing the B.1.351 variant.
When faced against the B.1.526 variant with the S477N mutation, they found high neutralizing titers comparable with the D614G. Although the neutralizing titers with E484K B.1.526 were lower by 3.6-fold. Both the B.1.526 and B.1351 variants have the E484K mutations, which the researchers note coincides with a similar decrease in neutralizing titers. Neutralizing activity with the Moderna vaccine showed comparable results.
Monoclonal antibody therapy versus B.1.526
The monoclonal antibodies neutralized B.1.526 variants with no titer loss and neutralized the B.1.526 with the S477N mutation with a high titer. Although, the therapy was 12-fold less active when exposed to the B.1.526 with the E484K mutation. "The cocktail which forms the REGN COV2 therapy potently neutralized the B.1.526 spike variants despite the partial loss of neutralizing activity against the E484K version of B.1.526."
Based on the findings, the B.1.526 variant is highly unlikely to evade neutralizing antibodies produced from either the vaccine or monoclonal antibody therapy.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources