Arizona joins the ever-growing list of American states harboring a variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A study led by Efrem S. Lim of Arizona State University confirms evidence of the B.1.243.1 with an E484K mutation on the spike protein.
B.1.427 and B.1.429 were first reported in California, and recent work has shown an emergence of the B.1.525 and B.1.526 variants in New York. This is in addition to other circulating coronavirus variants that were first detected in Brazil, the United Kingdom, and South Africa.
The researchers write:
"Based on the mutation profile, regional introduction and phylogenetic evidence, we recommend vigilant monitoring of B.1.243.1 as a potential variant of interest. This study demonstrates the need for sustained genomic surveillance in public health strategies and responses."
The study "Emergence of a SARS-CoV-2 E484K variant of interest in Arizona" is available as a preprint on the medRxiv* server, while the article undergoes peer review.
Tracking down viral variants
The team collected saliva samples from several Arizona counties that tested positive for SARs-CoV-2. They extracted RNA from 688 samples and performed genomic surveillance to look for variants. About 92.7% of genomes were successfully sequenced, where 7.7% had the B.1.1.7 variant, 33.5% had the B.1.427/429 variant, and 0.9% had the P.2 variant. A significant limitation in the study is that they did not sequence the entire sample as most genomes had low viral loads with high CTs in the 30s.
There was a total of 7 genomes with the B.1.243 variant and an E484K mutation. This new variant had 11 other unique mutations separate from mutations observed in the original B.1.243 lineage with the D614G mutation. This included V213G and E484K in the spike protein, a 9-nt deletion in ORF1ab, and a 3- nt insertion in the non-coding intergenic region upstream of the N gene, as well as other substitutions.
Based on the mutational profile, the researchers gave this E484K harboring variant the provisional name of B.1.243.1.
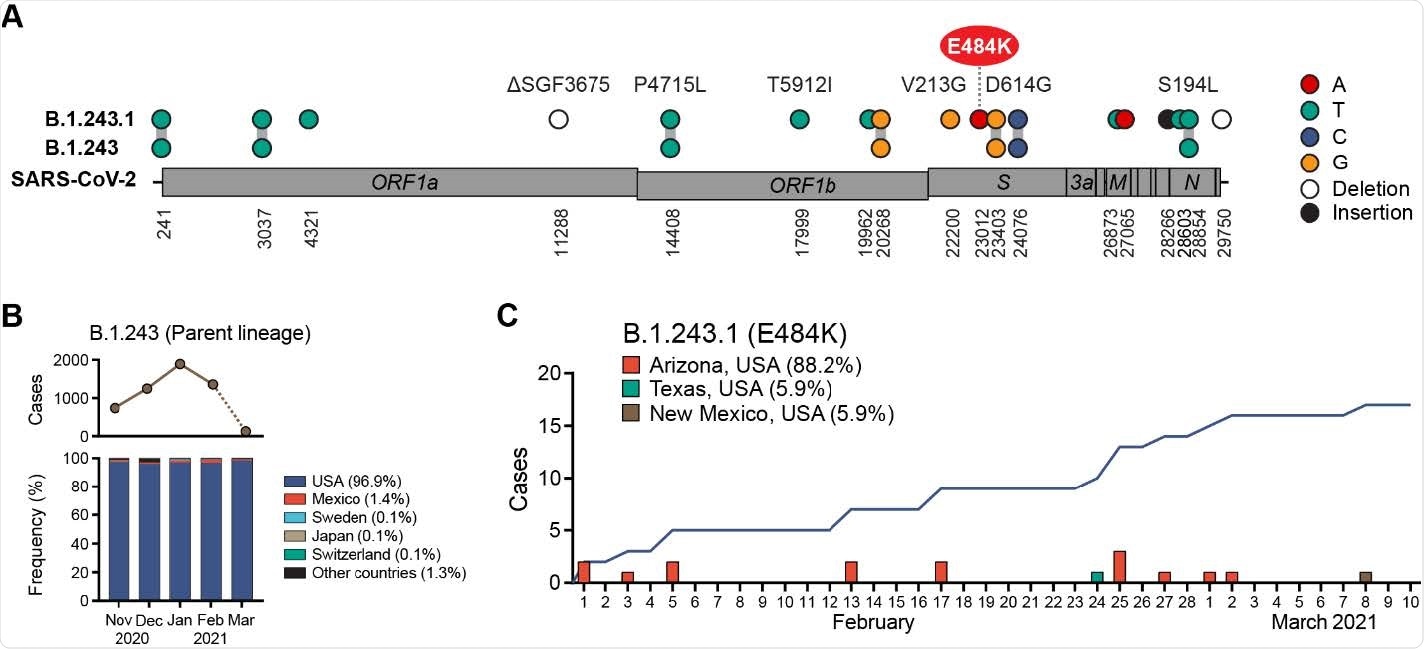
Emergence of E484K harboring B.1.243.1 variant in Arizona, USA. (A) B.1.243.1 lineage-defining mutations are shown on the SARS-CoV-2 genome. Mutations are shown in reference to the SARS-CoV-2 Wuhan-1 genome position (NC_045512.2). (B) Global prevalence of B.1.243 parental lineage from November 2021 to March 2021. Monthly number of B.1.243 cases (top) and distribution by country (bottom) are shown. Average frequency for each country is shown in parenthesis. Sequences from March are under-reported at the time of this reporting (indicated by dashed line). (C) B.1.243.1 case incidence reporting from February to March 2021. Cumulative case incidence is plotted as a line graph.
B.1.243 variant crosses state lines
Using the GISAID public database, the researchers looked at the geospatial distribution of this new variant. They found the B.1.243 variant and the E484K mutation has only recently emerged in Arizona, with 15 of the 17 sequenced cases occurring from February 1 to March 2.
One case of the B.1.243 variant and the E484K mutation was reported in Houston, Texas, on February 24 and another more recently in New Mexico, suggesting the variant is not contained in Arizona.
There were also two instances where the B.1.243 lineages independently attained the E484K mutation. Although, this variant lacked the 11 mutations observed in the new variant and appeared to be dead-end transmission events.
The researchers traced the variant to a monophyletic clade within the 20A/B.1.243 clade. And continued branching from the variant indicates the lineage continues to diversify. "This suggests that B.1.243.1 is being established in circulation within Arizona. In contrast, the two B.1.243 cases bearing the E484K mutation alone were phylogenetically distinct from the B.1.243.1 clade, supporting that they had evolved independently," write the research team.
Looking ahead
While the variant is starting to move to other states, human actions such as deciding to wear a mask and social distancing, and public policy measures can influence transmission rate. They suggest contact tracing and increases in genomic surveillance are necessary to measure the true extent of the risk of spreading this potential variant of interest.
"Genomic, epidemiologic and phylogenetic evidence indicates that the B.1.243.1 variant of interest is poised to emerge. These findings demonstrate the critical need to continue tracking SARS-CoV-2 in real-time to inform public health strategies, diagnostics, medical countermeasures and vaccines."
Important Notice
*medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.