New research led by Costas D. Maranas from The Pennsylvania State University predicts amino acid changes to the receptor-binding domain of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein would negatively impact binding affinity and subsequent infection into human cells.
Their results were derived from a novel two-step procedure called neural network molecular mechanics/Poisson-Boltzmann surface area (NN_MM-GBSA) that calculated binding energy from receptor-binding domain variants to human angiotensin-converting enzyme 2 (ACE2) receptors. The second step would construct a neural network from the findings to predict binding affinity. The team achieved an 82.2% accuracy rate for categorizing amino acid substitutions as helpful or unhelpful in a variant's binding affinity.
The researchers write:
"Our method thus sets up a framework for effectively screening binding affinity change with unknown single and multiple amino-acid changes. This can be a very valuable tool to predict host adaptation and zoonotic spillover of current and future SARS-CoV-2 variants."
The study "Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike protein and the human ACE2 receptor" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
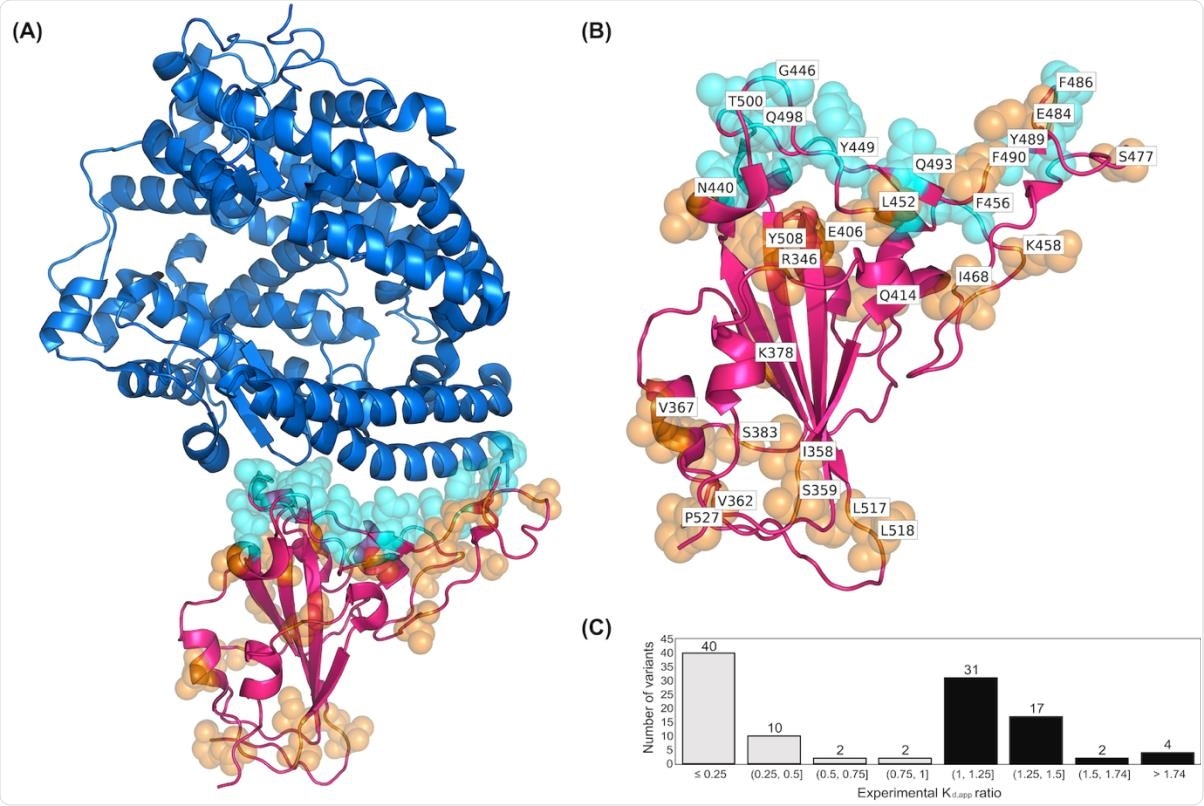
(A) The crystal structure of complex formed between RBD and hACE2 complex. The ACE2 protein is shown as a surface representation in blue and the RBD is shown in magenta. (B) Residues of the RBD variants that are in direct contact with hACE2 are depicted as cyan spheres. Residues that are not in direct contact are orange. (C) Histogramshowing experimental K D,app ratios for all 108 RBD variants in the dataset. The histogram bars in black denote number of variants in the training set with increasing binding affinity compared to WT (K D,app ratio>1.0) and the bars in gray indicate the variant counts with decreasing binding affinity (K D,app ratio < 1.0).
Study results
The team used a set of 108 variants to evaluate the binding energy and affinity of the receptor-binding domain of coronavirus variants. The predictions were based on MM-GBSA binding energies with a partial agreement with experimental data. Afterward, the binding energy was used to train a neural network regression model using decomposed MM-GBSA energy terms and dissociation constants ratio amongst coronavirus variants and the original wild-type strain.
"The study reported apparent dissociation constant KD,app ratios for all possible variants with single-amino acid changes at every RBD position. A KD,app ratio (i.e., KD,app,WT/KD,app,variant) for a variant greater than one implies stronger binding compared to WT, whereas a value less than one implies weaker binding."
Schematic representation of the workflow for building NN_MM-GBSA model. (A) MD simulations were performed for each single point amino acid substitution variant in explicit solvent followed by MM-GBSA analysis to calculate the decomposed components of binding energies. (B) MM-GBSA binding energy components were fed as inputs to the Neural network with the experimental Kd,app ratios as the regression target. The model is trained using five cycles of the five-fold cross-validation procedure.
Their model showed robust accuracy in their results with a 0.69 Pearson correlation coefficient between predicted and experimental values and an 82% accurate prediction for an amino acid substitution and its effect on binding. They observed no trendline to the training dataset as they continued to add more variants.
The weakest binding affinities appeared to occur when single amino acid substitutions were far-off from the spike protein's receptor-binding domain.
The amino acid changes in the E484K, N501Y, and K417N mutations from the South African B.1.351 variant and the E484K, N501Y, and K417T mutations in the Brazilian P1 variant increased the binding affinity for human ACE2 receptors. The researchers note K417N and K417T are known to decrease binding affinity. However, given that the variants became more infectious, the researchers suggest E484K and N501Y dominated the virus and exert a greater influence on binding affinity.
The team next looked at variants with double mutations, which have been suspected of being more infectious. They evaluated the binding affinity of two variants with either V503W and E406F or V503W and Y505W. The variants had large hydrophobic amino acids that contributed to a higher binding affinity with human ACE2 receptors. Hydrophobic interactions have mainly been associated with solid anchors for binding.
Limitations to NN_MM-GBSA
The researchers note a significant downside to the two-step procedure: it's heavily time-consuming and reliant on theoretical deductions on binding energy and subsequent affinity to human ACE2 receptors. While this limitation may be circumvented by using existing energy terms from the balanced training set of the 108 variants to make new predictions, it would require retraining the neural network model.
Future work using the two-step procedure could focus on coronavirus cases in animals. Previous research has shown evidence of SARS-CoV-2 infecting cats, dogs, and ferrets but how variants affect these species remains unknown. "Assuming that training of NN_MM-GBSA using hACE2 data is robust, it could in principle be used to prospectively assess, the relative affinity of the RBD of circulating variants for various animal ACE2s."
Important Notice
*bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.