Most research testing monoclonal antibodies specific to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) use antigen probes from plasmablasts and memory B cells from donated plasma of recovered SARS-CoV-2 individuals. However, given the rise of new variants of concern worldwide, the motivation to produce more antibody-based tools against SARS-CoV-2 is high.
How they did it
The researchers collected monoclonal antibodies specific to SARS-CoV-2 from plasmablasts and memory B cells of 126 patients with COVID-19 infection. The samples were collected in April 2020 in New York City, indicating the immune response reflects the first wave of the pandemic. They collected. A total of 119 antibodies targeted the SARS-CoV-2 spike protein, 106 targeted the receptor-binding domain, and 122 targeted the N-terminal domain.
The plasma was tested for neutralizing potential whether a wide range of neutralizing titers from <40 to 765 were detected. The potency was weakly correlated with antibody levels for the spike protein, receptor binding domain, and N-terminal domain. Also, some plasma samples had no neutralizing ability despite high antibody levels.
To develop monoclonal antibodies with high specificity for SARS-CoV-2 epitopes, the researchers used a single-cell assay to screen for antibodies in plasmablasts and memory B cells. Assays extracted a total of 169 monoclonal antibodies targeting SARS-CoV-2 from plasmablasts and 47 from memory B cells of 12 individuals. About 59 of the monoclonal antibodies target the receptor-binding domain, 64 targeted the N-terminal domain, and 46 targeted neither, suggesting antibodies can detect the entire spike protein.
Plasmablasts and memory B cells generate highly effective neutralizing antibodies
The researchers found that most monoclonal antibodies had no neutralizing potency, but 21 antibodies showed strong potency against SARS-CoV-2.
Antibodies of the immunoglobulin A (IgA) isotype demonstrated superior neutralization. Twenty-one antibodies filed under the immunoglobulin G (IgG) isotype targeted several spike protein areas — 16 targeted the receptor-binding domain, and 5 targeted the N-terminal domain.
The average potency from antibodies generated from plasmablasts or memory B cells were similar, indicating they both produce potent antibodies.
Using a high-throughput surface plasmon resonance, antibodies specific to the receptor-binding domain showed greater affinity than N-terminal domain-specific monoclonal antibodies. The N-terminal domain-specific monoclonal antibodies from plasmablasts had lower affinities than antibodies from memory B cells.
The researchers found the most potent monoclonal antibody had a low affinity for the N-terminal domain.
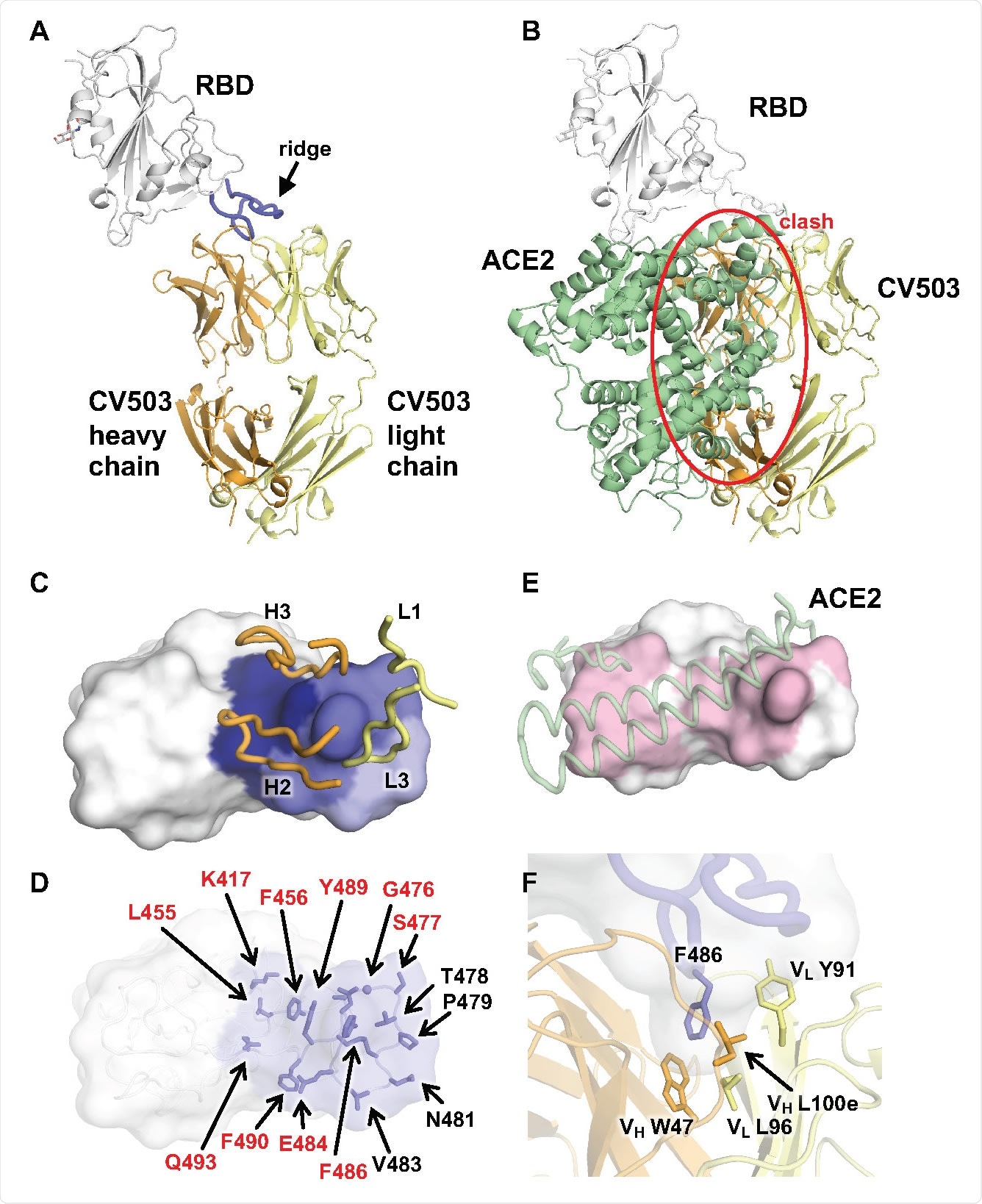
Crystal structure of SARS-CoV-2 RBD in complex with CV503. A, CV503 binds to the ridge region of SARS-CoV-2 RBD. The heavy and light chains of CV503 are shown in orange and yellow, respectively. SARS-CoV-2 RBD is in white, where its ridge region (residues 471–491) is shown in blue. B, The ACE2/RBD complex structure (PDB ID: 6M0J)52 is superimposed on the CV503/RBD complex. The heavy chain of CV503 (orange) would clash with ACE2 (green) if bound to RBD simultaneously (indicated by red circle). C-D, Epitope of CV503. Epitope residues contacting the heavy chain are in dark blue and light chain are in light blue, while residues contacting both heavy and light chains are in ocean blue. In C, CDR loops that are directly involved in RBD-binding are labeled. In D, epitope residues are labeled. Epitope residues that are also involved in ACE2 binding are labeled in red. E, ACE2-binding site on the RBD are in light pink. ACE2 is represented as semitransparent cartoon in pale green. Epitope residues and ACE2-interacting residues are defined as those with a buried surface area (BSA) > 0 Å2. F, F486 at the ridge region of SARS-CoV-2 RBD (blue) is clamped in a hydrophobic pocket formed by the heavy (orange) and light chains (yellow) of CV503.
Antibody binding ability to COVID-19 variants
About 28 out of 37 monoclonal antibodies specific to the receptor-binding domain and 3 out of 20 antibodies specific to the N-terminal domain had over a 90% binding to the B.1.1.7 variant.
Only 10 antibodies specific to the receptor-binding domain and 2 antibodies specific to the N-terminal domain retained full binding to the B.1.351 variant. The researchers suggest the results provide more evidence the B.1.351 variant is more successful in the immune escape.
Bispecific antibodies on SARS-CoV-2
Considering most monoclonal antibodies bind to non-overlapping sites, they then tested for possible synergy between antibodies. They merged antibody specificities to produce ten bispecific antibodies combining variable regions of potent neutralizers with two types of linkers.
The bispecific antibodies that could target the receptor-binding domain and the N-terminal domain retained binding to both domains. When testing bispecific antibodies in SARS-CoV-2 assays, five antibodies — CV503_521_GS, CV521_1182_GS, CV1206_521_GS, CV521_503_GS, and CV503_664_EL neutralized SARS-CoV-2.
"Notably, CV1206_521_GS neutralizes SARS-CoV-2 with >100-fold higher potency than a cocktail of its constituent antibodies CV1206 and CV521. We found that CV1206_521_GS uses its inner and outer Fab domains to cross-link NTD and RBD in adjacent spike proteins, a mode of action that is unavailable to conventional mAbs, even when used in combination."
The researchers suggest results appear promising that combining bispecific antibodies could further enhance neutralizing potential.
When tested against the B.1.1.7 and B.1.351 variants' spike protein, only bispecific antibodies whose two components lost binding to variants were unable to bind. This suggests this antibody form is more resistant to spike mutations than typical monoclonal antibodies.
All bispecific antibodies neutralized the D614G strain, most bispecific antibodies with CV521 neutralized the B.1.1.7 variant, and six out of nine bispecific antibodies neutralized B.1.351.
Important Notice
*bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.