The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified in Wuhan, China, in late December 2019. The virus triggers multiple clinical complications in humans and can lead to COVID-19 (coronavirus disease 2019). To date, SARS-CoV-2 has infected over 132 million individuals and caused over 2.88 million deaths worldwide.
COVID-19 is a heterogeneous disease, primarily characterized by fever, cough and acute respiratory distress syndrome (ARDS), and in-hospital mortality. Severe COVID-19 is associated with comorbidities such as cardiovascular disease, hypertension, and diabetes mellitus and coagulopathy.
Coagulopathy - a condition in which the blood's ability to clot is impaired, is recognized as a significant aspect of morbidity in COVID-19 patients. In a recent study, Prof Lewis Cantley and a team of researchers explored the role of coagulation factors in the SARS-CoV-2 infection. Their recent study is posted on the bioRxiv* server.
"Coagulation-induced cleavage enhances spike activation and increases viral entry into target cells."
They found that the proteases involved in clotting can also directly cleave the SARS-CoV-2 glycoprotein spike, enhancing the viral entry. They also found that protease inhibitors promiscuously inhibited the spike cleavage by both transmembrane serine proteases as well as coagulation factors. Spike cleavage by proteases is crucial to the viral entry; it facilitates membrane fusion and hijacks of the host cell by the viral genome.
The spike protein consists of S1 and S2 subunits. The host cell proteases act on the S1/S2 subunit boundary and at the S2' region proximal to the fusion peptide. This cleavage opens up the spike trimer and exposes the S2' site, which must be cleaved to allow for the release of the fusion peptide. One of the promising antiviral drug targets for SARS-CoV-2 is the TMPRSS2 which is an important host cell factor in the proteolytic activation across multiple coronaviruses.
Camostat and Nafamostat are two broad-spectrum inhibitors and act against TMPRSS2. While these two drugs are clinically (approved for other applications) in Japan, these are not currently FDA-approved for any indication in the US. In this study, the researchers found that Nafamostat broadly inhibits the cleavage of spike peptides by both transmembrane serine proteases and the coagulation factors.
Even as anti-SARS-CoV-2 vaccines are currently administered across the world, the continuing existence of a large reservoir of active cases over a long period will cause viral variants to emerge.
Pathogenesis of SARS-CoV-2 infection is associated with coagulopathy and thromboembolic events. Signs of dysregulated blood clotting were evident in SARS-CoV-2 infected patients who were admitted to hospitals. These were 1) elevated levels of D-dimer levels (an indicator of fibrinolysis and coagulopathy), 2) low platelet counts (an indicator of consumptive coagulopathy), 3) elevated systemic activity of clotting factors V, VIII, and X.
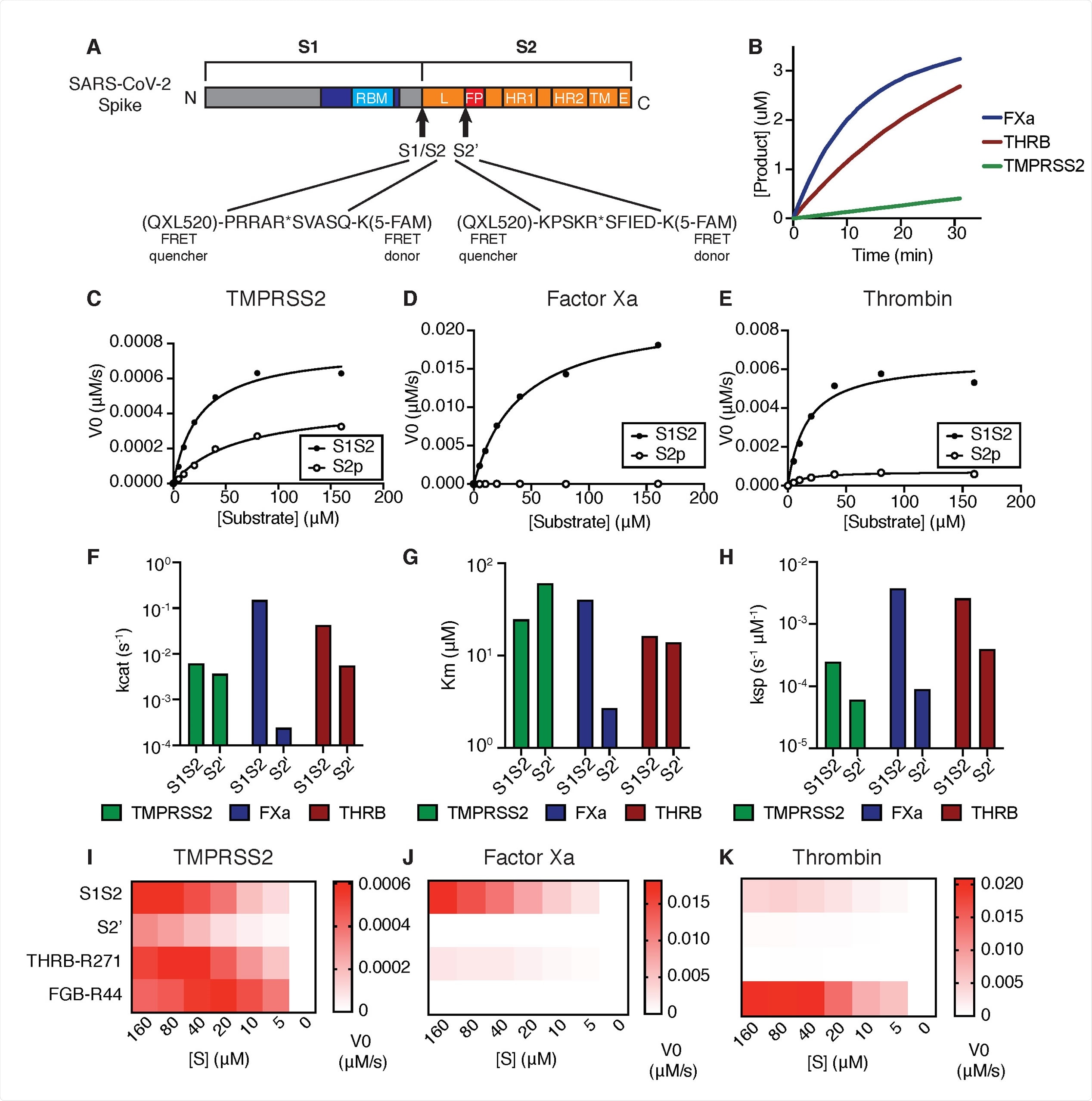
Coagulation factors directly cleave SARS-CoV-2 spike. (A) Peptides derived from two known cleavage sites of SARS-CoV-2 spike were designed with C-terminal fluorophore 5-FAM and N-terminal FRET quencher QXL-520. (B) Cleavage of 10 μM S1/S2 peptide substrate incubated with 125nM TMPRSS2, factor Xa, or thrombin. Initial velocities for the cleavage of SARS-CoV-2 spike S1/S2 and S2’ peptide substrates by (C) TMPRSS2, (D) Factor Xa, and (E) Thrombin were measured over a range of 0-160 μM substrate. From initial velocity values, enzyme kinetic constants (F) turnover rate Kcat (s-1), (G) affinity constant Km, and (H) specificity constant (Kcat/Km) were obtained for the indicated enzymes with S1/S2 and S2’ peptides. (I-K) Heatmaps depict the initial velocity V0 of cleavage of the indicated peptide substrates and concentrations by (I) TMPRSS2, (J) factor Xa, and (K) thrombin.
Because the coagulation cascade is orchestrated by a chain reaction of serine protease zymogens that are each activated by proteolytic processing, the researchers investigated if the proteases involved in the clotting also play a role in the SARS-CoV-2 spike cleavage.
They found that the coagulation factors, serine proteases factor Xa and thrombin, directly cleave SARS-CoV-2 spike, with more significant proteolytic activity against the SARS-CoV-2 peptide substrates than TMPRSS2. They also found that the S1/S2 boundary appears to be an even more optimal factor Xa substrate than peptide substrates derived from known physiological targets of factor Xa in coagulation.
They reported that the serine protease inhibitors suppress SARS-CoV-2 entry via inhibition of TMPRSS2. They observed that the TMPRSS2 is likely the primary cellular target of camostat and nafamostat.
Further, they found that the nafamostat is a versatile inhibitor of spike activation by various transmembrane serine proteases (TTSPs) and coagulation factors.
"We have mitigated the risk of artifact by using multiple orthogonal platforms, including VSV-based pseudovirus, HIV-based pseudovirus, and syncytia formation."
The researchers detailed the clinical relevance of potential antiviral activity of anticoagulants, bleeding risks and the limitations in this study.
COVID-19 associated events (such as acute lung injury from viral cytopathic effects, cytokine storm, complement activation, and antiphospholipid autoantibodies) are known to instigate a coagulation cascade. While the coordination between inflammation and hemostasis is well understood, the researchers in this study point out that the precise molecular mechanism connecting the SARS-CoV-2 infection and dysregulation of the hemostasis is yet unclear.
In this study, Using a FRET-based enzymatic assay and multiple platforms of pseudovirus and cell
fusion assays, the researchers investigated and confirmed that the circulating proteases involved in blood clotting contribute to SARS-CoV-2 spike cleavage activation and thus, enhance the viral entry. Based on their study, the researchers proposed that the nafamostat, among the currently available drugs, is best suited as a multi-purpose inhibitor against spike cleavage by TTSPs and coagulation factors.
This study provides relevant data to explore an early intervention with judiciously selected anticoagulant treatment for SARS-CoV-2 infection. Preparedness to mitigate a future SARS-CoV-3 epidemic is critical to pursue through the understanding of coronavirus-host interactions, the researchers emphasize.
"Perhaps, SARS-CoV-2 has undergone selection to both induce and exploit an environment locally enriched in coagulation proteases, instigating a positive feedback loop to promote entry into additional host cells."
Journal reference:
- Coagulation factors directly cleave SARS-CoV-2 spike and enhance viral entry, Edward R. Kastenhuber, Javier A. Jaimes, Jared L. Johnson, Marisa Mercadante, Frauke Muecksch, Yiska Weisblum, Yaron Bram, Robert E. Schwartz, Gary R. Whittaker, Lewis C. Cantley, bioRxiv 2021.03.31.437960; doi: https://doi.org/10.1101/2021.03.31.437960, https://www.biorxiv.org/content/10.1101/2021.03.31.437960v1