The causative agent of the coronavirus disease 2019 (COVID-19), SARS-CoV-2, belongs to the Betacoronavirus genus and Sarbecovirus subgenus of the Coronaviridae family, which is a group of enveloped, single-stranded RNA viruses.
Other representatives of Sarbecoviruses include SARS-CoV (which was the coronavirus that led to the SARS outbreak in 2002 and 2003), but also a myriad of SARS-like bat coronaviruses that have been identified and analyzed in depth.
Among the viral structural proteins, the spike glycoprotein has a pivotal role in the host range, cell tropism, and entrance, as well as infectivity traits. Hence, it is no wonder that it is considered a prime target of the host immune response.
Therefore, a thorough comparative analysis of viral proteins would help immensely in our better understanding of viral biology and pathology, providing insights into its origin, as well as the conditions that led to the ongoing COVID-19 pandemic.
This is why Dr. Matthew Cotten, Dr. David L. Roberston, and Dr. My V.T. Phan from Uganda and the United Kingdom aimed to identify unique peptide regions of SARS-CoV-2 when compared to all available Sarbecoviruses in order to appraise the features that might enable SARS-CoV-2 to replicate and transmit efficiently among us.
.jpg)
Study: Unique protein features of SARS-CoV-2 relative to other Sarbecoviruses. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Comparing genomes and appraising evolutionary distances
In a nutshell, this research group has explored the genomes across the Sarbecovirus subgenus by using profile hidden Markov models. This modeling approach is based on a statistical description of the properties of viral proteins and their amino acid sequences.
More specifically, ten early SARS-CoV-2 genomes were compared to a representative subset of Sarbecovirus genomes obtained from human individuals, bats, pangolins, and civet cats. These were selected after the same analysis has been applied to all available Betacoronavirus genomes in order to avoid missing any surprisingly close viral genome regions.
In order to appraise total domain distances between viral groups, normalized bit-score sums (grouped into SARS-CoV-2 and Sarbecoviruses from human, bat, pangolin and civet cat) were summed for all domains and for each genome.
Analysis scheme. (A) Profile Hidden Markov Model (pHMM) domains were generated from a set of 35 early lineage B SARS-CoV-2 genome sequences. All open reading frames were translated and then sliced into either 44 amino acid peptides with a step size of 22 amino acids or 15 amino acid peptides with a step size of 8 amino acid. The peptides were clustered using Uclust (13), aligned with MAFFT (14) and then each alignment was built into a pHMM using HMMER-3 (10). (B) The set of pHMMs were used to query Sarbecovirus genome sequences, bit scores were collected as a measure of similarity between each pHMM and the query sequence. (C) Bit-scores were gathered an analyzed to detect regions that differ between early SARS-CoV-2 genomes and query genomes.
Unique nature of SARS-CoV-2
Detected changes in spike glycoprotein of SARS-CoV-2 – in comparison to a large set of known Sarbecovirus – reveal that the recent zoonotic source of this virus is yet to be found but also underpin a rather unique nature of the SARS-CoV-2 genome.
In line with previous reports, a small set of bat and pangolin-derived Sarbecoviruses demonstrate the most significant similarity to SARS-CoV-2, while a measure of proteome similarity showed that it is unlikely that bat Sarbecoviruses are the direct source of the pandemic virus.
Furthermore, the regions of variance that were identified in this study may indicate either functional changes in SARS-CoV-2 proteins or amino acid positions that can be modified without compromising the requisite functions of the protein.
The detailed spike analysis revealed 82 domains of 15 amino acids that have demonstrated high variation in the Sarbecoviruses, while 29 of these domains show changes in variants of concern relative to the early lineage of SARS-CoV-2.
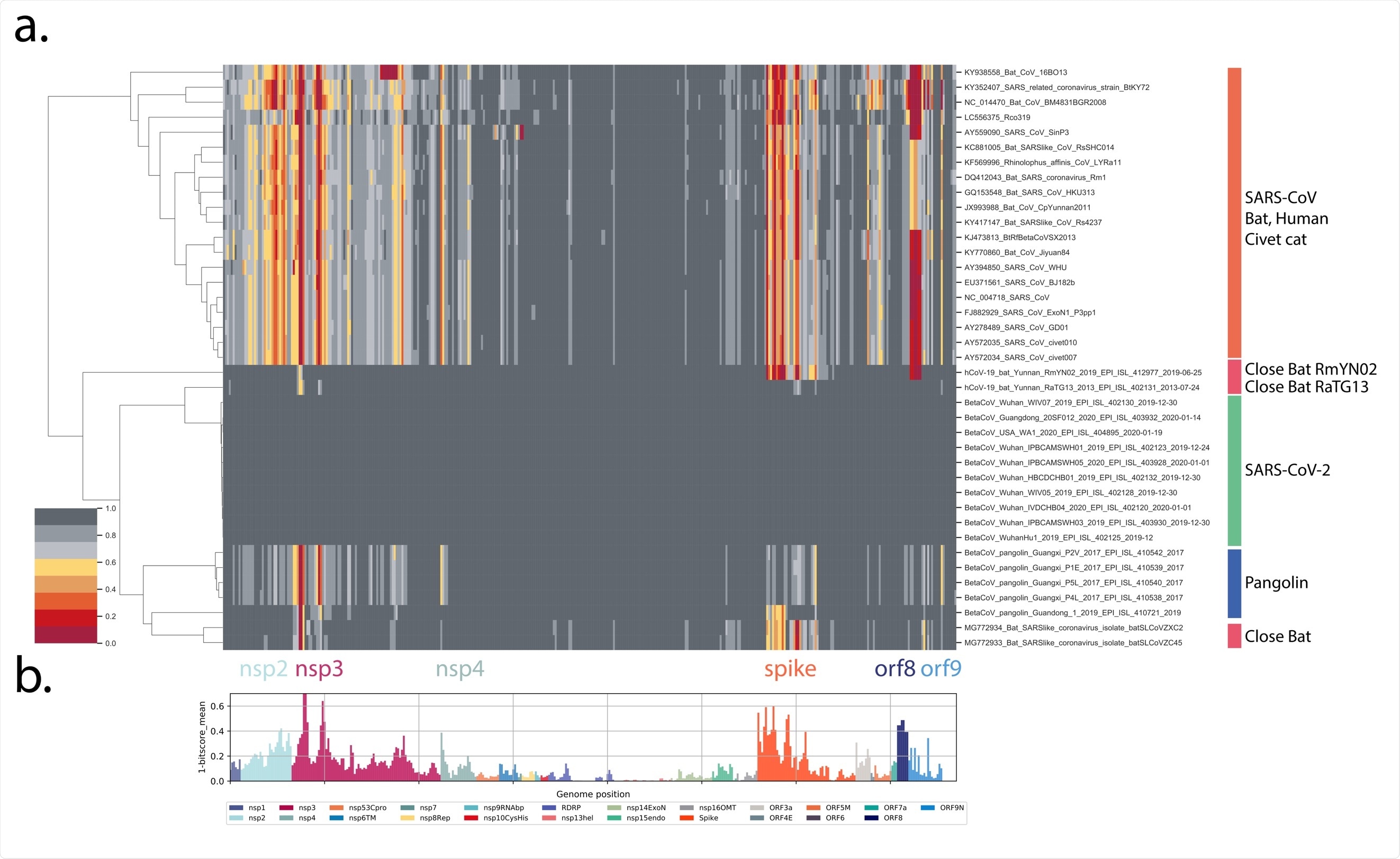
Proteome differences in SARS-CoV-2 vs close Bat, Human and Civet cat Sarbecoviruses. All forward open reading frames from the 35 early lineage B SARS-CoV-2 genomes were translated, and processed into 44 aa peptides (with 22 aa overlap), clustered at 0.65 identity using Uclust (11), aligned with MAAFT (12) and converted into pHMMs using HMMER-3 (10). The presence of these domains was sought in a set of Sarbecovirus genomes plus the SARS-CoV-2 genomes and genomes were then clustered using hierarchical clustering based on the normalized domain bit-scores (e.g. the similarity of the identified query domain to the reference lineage B SARS-CoV-2 domain). Each row represents a genome, each column represents a domain. Domains are displayed in their order across the SARS-CoV-2 genome, Red = low normalized domain bit-score (lower similarity to lineage B SARS-CoV-2) = distant from SARS-CoV-2, Darkest grey = normalized domain bit-score = 1 = highly similar to lineage B SARS-CoV-2. Groups of coronaviruses were indicated to the right of the figure. (A) Domain differences across the Sarbecovirus subgenus. (B) For each domain the mean bit-score was calculated across the entire set of Sarbecovirus genomes and the value 1-mean bit-score was plotted for each domain. Domains are colored by the proteins from which they were derived with the color code indicated below the figure.
Being wary of the viral adaptation
This study gives credence to the notion of continuous genomic variant surveillance, which should then translate to the preparedness of vaccine producers to accommodate such spike glycoprotein changes in the next generation of vaccine updates.
"In broad terms, the SARS-CoV-2 evolution observed in the current variants of concern has sampled only 36% of the possible spikes changes which have occurred historically in Sarbecovirus evolution", say study authors in this bioRxiv paper.
"It is highly likely that a large number of new SARS-CoV-2 variants with changes in these regions are possible, compatible with virus replication and expected in the coming months unless global viral replication is severely reduced", they add.
In conclusion, such a high mutation rate of SARS-CoV-2 in combination with the remarkable number of SARS-CoV-2 infections worldwide gives rise to massive viral adaptation. Hence, further experiments will be necessary to discern true functional changes from neutral evolution.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources