The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome generates 16 distinct non-structural proteins (nsp) that constitute the enzymes and accessory proteins responsible for virus replication once inside a host cell. These protein complexes produce and cap RNA strands that will go on to be translated by the host machinery. Capping the RNA in a manner that is recognized as similar to endogenous mRNA ensures compatibility, lessens RNA degradation rates and lowers the probability of triggering an immune response in the host cell.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The function of nsp14
Following synthesis of the viral RNA strand, nsp13 is involved in preparing the strand for capping by removing the terminal phosphate group, after which nsp12 transfers the GpppA cap structure to the 5’ end of the RNA. Nsp14, nsp16, and nsp10 then finalize the cap structure by adding several methyl groups, forming Cap1.
The group demonstrated that methylation performed by the nsp16/nsp10 complex is dependent upon the prior methylation of the strand by nsp14, and both of these enzymes require the presence of S-adenosyl-L-methionine (SAM), a methyl donor, producing S-adenosyl-L-homocysteine (SAH) upon donation. Nsp14 was shown to be a guanine-N7 methyltransferase that can only catalyze the methylation of the starting GpppA structure.
Nsp14 was purified and the group developed an assay to quantifiably determine the conversion of SAM to SAH in the presence of the enzyme and GppA-capped RNA, thereby determining the activity of the methyltransferase enzyme. Nsp14 was also screened against a library of over 5,000 chemical compounds in search of antiviral drug leads, applied at a concentration of 3.125 μM, and each ranked based on efficacy. The resulting drug leads were further refined by removing probable screening errors, compounds that would be difficult to source, or those with known toxicities at the indicated required concentration.
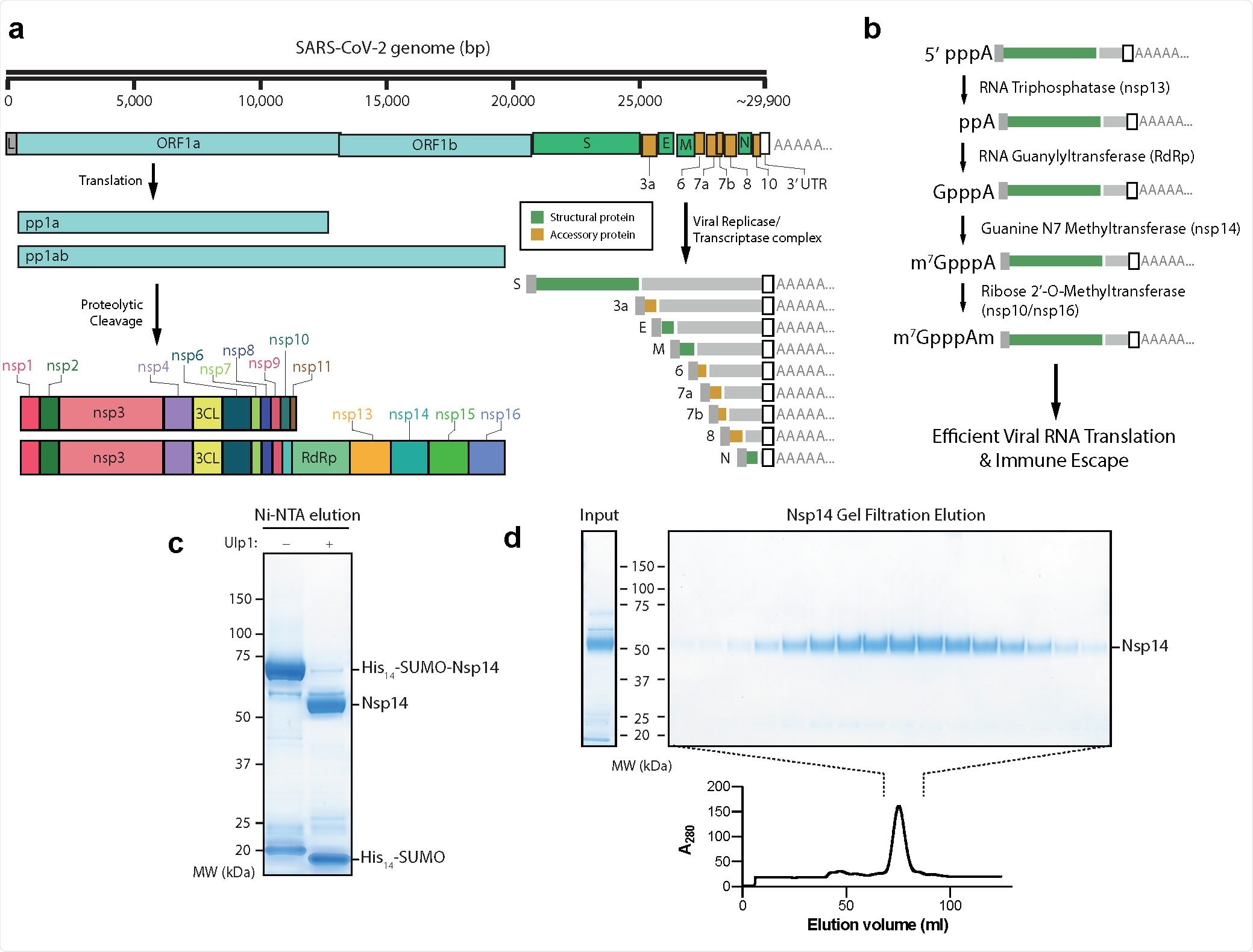
Purification of nsp14 Guanine N7 Methyltransferase a) Outline of the SARS-CoV-2 genome. Pp1a and Pp1ab represent polyproteins a and ab, respectively. Pp1a and pp1ab are able to autoproteolytically cleave themselves to form the nsp proteins outlined. The viral replicase/transcriptase complex produces a series of nested viral RNAs that encode accessory (orange) or structural (green) viral proteins. b) Viral RNA capping outline. The initial RNA nucleotide possesses a γ and β phosphate, unlike following RNA bases. The γ phosphate is removed by nsp13, followed by the addition of Gp by nsp12, releasing pyrophosphate. nsp14 and nsp16/10 then catalyze the formation of the final Cap-0 structure. c) Coomassie gel of His14-SUMO cleavage. Left column: Elution from Ni-NTA beads without the Ulp1 SUMO-dependent protease. Right: Elution from Ni-NTA beads after treatment with Ulp1 (see Methods). d) Gel filtration fractions of nsp14. Left: Input to gel filtration. Right: Pooled fractions from the main peak of the elution (lower). nsp14 expected size: 55 kDa.
Nsp14 inhibitors
A chemical compound termed PF-03882845 was found to be the most potent inhibitor, exhibiting an inhibitory concentration 50% of 1.1 μM. Trifluperidol, Inauhzin, and Lomeguatrib had IC50 values of 12.9 μM, 23 μM, and 59.8 μM, respectively. The activity of these drugs towards the nsp16/nsp10 complex was also tested, and none were found to inhibit the complex, demonstrating the specificity of the drugs towards nsp14.
Mammalian cells were then infected with SARS-CoV-2, and the inhibitors applied, followed by fluorescent staining of anti-nucleoprotein antibodies, allowing the group to track both viral load and cytotoxicity towards the cells. PF-03882845, Inauhzin, and Trifluperidol each had IC50 values of between 11-13 μM, in order of lessening potency, while Lomeguatrib required a concentration of almost 60 μM for similar inhibition levels. None of the drugs proved to be cytotoxic in the cell line tested.
Antiviral drug remdesivir generally requires concentrations of around 1 μM to lower viral load in the cells tested by the group. To test synergy, the drug was applied at a concentration of only 0.5 μM in combination with one of the four lead drugs. In this case, Inauhzin exhibited similar IC50 values when applied alone, while the three other drugs exhibited significant synergy, having notably lower IC50 values. The IC50 of PF-03882845 and Trifluperidol lowered in particular, dropping to 4.79 μM and 5.05 μM, respectively.
PF-03882845 and Inauhzin have previously been in clinical trials as a mineralocorticoid receptor agonist and SIRT inhibitor, respectively, though neither has yet been approved for human use. Trifluperidol is a licensed therapeutic used to treat various psychoses but has been implicated with various detrimental side effects under long-term use, and so may not make an ideal COVID-19 prophylactic. In any case, these drugs may contribute to the development of more effective SARS-CoV-2 drugs.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Identification of SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of the nsp14 RNA Cap Methyltransferase, Souradeep Basu, Tiffany Mak, Rachel Ulferts, Mary Wu, Tom Deegan, Ryo Fujisawa, Kang Wei Tan, Chew Theng Lim, Clovis Basier, Berta Canal, Joseph F. Curran, Lucy Drury, Allison W. McClure, Emma L. Roberts, Florian Weissmann, Theresa U. Zeisner, Rupert Beale, Victoria H. Cowling, Michael Howell, Karim Labib, John F.X. Diffley, bioRxiv, 2021.04.07.438810; doi: https://doi.org/10.1101/2021.04.07.438810, https://www.biorxiv.org/content/10.1101/2021.04.07.438810v1
- Peer reviewed and published scientific report.
Basu, Souradeep, Tiffany Mak, Rachel Ulferts, Mary Wu, Tom Deegan, Ryo Fujisawa, Kang Wei Tan, et al. 2021. “Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of Nsp14 RNA Cap Methyltransferase.” Biochemical Journal 478 (13): 2481–97. https://doi.org/10.1042/bcj20210219. https://portlandpress.com/biochemj/article/478/13/2481/229153/Identifying-SARS-CoV-2-antiviral-compounds-by.