The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein is a class I fusion glycoprotein responsible for interacting with the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of human cells, enabling cell entry. It is the primary target of both natural and vaccine-acquired neutralizing antibodies.
Recent studies have suggested that the occupancy and structure of the spike protein glycan shield play a prominent role in host cell entry and immune function and that various factors can adjust the rate and order in which glycans coat the protein, potentially resulting in differing glycoforms.
Multiple N-glycans are strategically positioned around the opening of the receptor-binding domain to prevent water molecules from entering the cleft when in the open position, potentially weakening spike protein structure and affinity towards ACE2.
Specifically, the N-glycan at position N234 fills the cavity and supports the open conformation of the protein, while N-glycans at positions N165 and N343 have been implicated to be involved in stabilizing the open conformation and mediating the transition between states. These functions are facilitated by contacts made with adjacent protein residues. Thus, altering the sequence, length, and branching of the glycans at these sites influences the spike protein's conformational fluctuation and stability, potentially altering the affinity of SARS-CoV-2 towards the ACE2 receptor.
In a research paper recently uploaded to the preprint server bioRxiv* by Harbison et al. (April 1st 2021) a set of multi-microsecond long molecular dynamics simulations study the effects of altering glycosylation at sites N234, N165, N343, and the newly identified ancestral site N370, uncovering the influence of these sites on infectivity.
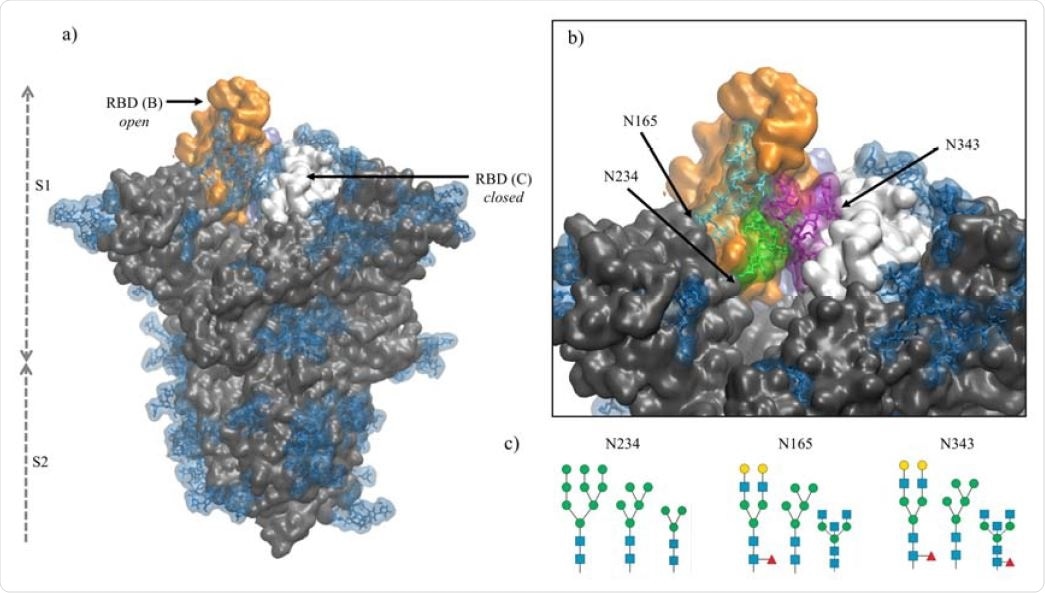
Panel a) Structure of the fully glycosylated SARS-CoV-2 S (PDBid 6VYB)6 ectodomain. The protein is shown in grey with the RBDs of chain B and C highlighted in orange and white, respectively; the glycan shield is highlighted in blue. Panel b) Close up on the open pockets with the N-glycans at the strategic positions N234, N165, and N343 highlighted in green, cyan and purple, respectively. Panel c) N-glycans considered in the different models studied in this work, represented through the SNFG23 and drawn with DrawGlycan24 (http://www.virtualglycome.org/DrawGlycan/). Molecular rendering done with VMD25.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Conformational stability and ACE2 affinity
The group began by replacing the N234 glycan with a shorter oligomannose structure (Man5), noting that when in the open state, the cavity of the receptor-binding domain was no longer as completely occupied, and the conformational flexibility was somewhat greater, though the open conformation was still considered stable. A slightly longer and more branched glycan (Man9) was also tested in this position and was able to fill the space more effectively and facilitate greater interaction with proteins on the opposite side of the cleft, further stabilizing the open conformation. Replacing glycans N165 and N343 with the shorter glycan, Man5, was noted to lessen conformational stability of the receptor-binding domain in the open position, but that the presence of the larger Man9 at position N234 was able to stabilize the open position sufficiently by occupying the open cleft. As the open conformation was relatively stable when replacing N234 with Man5 or Man9 the affinity towards the ACE2 receptor was maintained.
Glycan N234 was then replaced with a shorter oligomannose still (Man3), and the group notes that at this length, the glycan is unable to form functional interactions with cleft-adjacent proteins that contribute to stability. In this case, N-glycans at N165 and N343 dominated with regards to conformational interactions, forcing the receptor-binding domain into an intermediate conformation, between fully open and closed. This change in conformation via reduced open-conformation stability resulted in lessened affinity towards the ACE2 receptor.
SARS-CoV-2 glycan ancestry
The group also performed ancestral sequence reconstruction of SARS spike protein sequences, finding that compared to recent ancestors, SARS-CoV-2 is missing a sequon (an amino acid chain that anchors an N-glycan) at position N370. The group added this position to the SARS-CoV-2 sequence and again ran molecular dynamics simulations with Man9 at position N234. The open conformation was found to be equal in stability and affinity towards ACE2 to the formerly tested Man9-N234 model. However, that the majority of the receptor-binding domain cleft is filled by N370 in this case, with Man9-N234 being pushed to the pocket entrance.
Similarly, when in the closed state N370 was found to thread the two halves of the spike protein receptor-binding domain together, improving stability. However, improving the stability of the closed state lessens the probability of interacting with the ACE2 receptor, and thus lowers the virulence of the virus. The group proposes that the loss of glycosylation at N370 contributed strongly to the enhanced virulence observed in SARS-CoV-2 over SARS-CoV-1. According to this ancestral analysis, the role of the glycan at position N234 could have evolved relatively recently in SARS-CoV-2, growing to substitute for the loss of N370. This replacement could have served the evolutionary role of lowering the energetic cost of the receptor-binding domain entering the open conformational state.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Fine-tuning the Spike: Role of the nature and topology of the glycan shield in the structure and dynamics of SARS-CoV-2 S Aoife M. Harbison, Carl A. Fogarty, Toan K. Phung, Akash Satheesan, Benjamin L. Schulz, Elisa Fadda, bioRxiv, 2021.04.01.438036; doi: https://doi.org/10.1101/2021.04.01.438036, https://www.biorxiv.org/content/10.1101/2021.04.01.438036v1
- Peer reviewed and published scientific report.
Harbison, Aoife M., Carl A. Fogarty, Toan K. Phung, Akash Satheesan, Benjamin L. Schulz, and Elisa Fadda. 2022. “Fine-Tuning the Spike: Role of the Nature and Topology of the Glycan Shield in the Structure and Dynamics of the SARS-CoV-2 S.” Chemical Science 13 (2): 386–95. https://doi.org/10.1039/d1sc04832e. https://pubs.rsc.org/en/content/articlelanding/2022/SC/D1SC04832E.