Researchers in the United States have identified 95 high-frequency mutations in the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that threaten to compromise the efficacy of current and future vaccines and antibody therapies for coronavirus disease 2019 (COVID-19).
Using machine learning and deep learning models, Guo-Wei Wei and colleagues from Michigan State University identified the mutations in the receptor-binding domain (RBD) of the spike protein that the virus relies on to infect host cells.
The team says that as well as threatening to reduce the effectiveness of monoclonal antibodies (mAbs) that have received emergency use authorization (EUA), the mutations could also compromise the effectiveness of mAbs currently undergoing evaluation in clinical trials.
The study also provides a general perspective regarding how mutations will affect the currently EUA-approved Pfizer-BioNTech and Moderna vaccines.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
SARS-CoV-2 S protein antibodies are secreted by B cells in aiming to compete with the host ACE2 for binding to the S protein RBD

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Variants increasingly threaten therapeutic approaches
The ongoing rollout of mass vaccination and the development of mAbs therapies are the two most promising approaches to battling the COVID-19 pandemic.
However, rapidly emerging SARS-CoV-2 variants are increasingly posing a threat to the effectiveness of existing vaccines and mAbs.
The initial stage of the SARS-CoV-2 infection process is mediated by the viral spike protein, which attaches to the host cell receptor angiotensin-converting enzyme 2 (ACE2) via its RBD.
The spike RBD is the primary target of neutralizing antibodies following natural infection.
The Pfizer-BioNTech and Moderna vaccines that have been granted EUA in many countries are designed to trigger antibodies that bind to the spike protein and prevent it from attaching to ACE2. Similarly, mAb therapies primarily target the spike protein.
However, variants are emerging worldwide that contain mutations in the spike RBD, some of which have been shown to exhibit resistance to neutralizing activity.
“RBD mutations that enhance the RBD binding to ACE2 and weaken the RBD binding to many antibodies pose potential threats to vaccines and antibody therapies,” writes Wei and the team.
More about the variants and their mutations
Studies have already demonstrated that the E484K mutations found in the spike RBD of the South African (B.1.351) and Brazilian (P.1) variants enable SARS-CoV-2 to escape immune responses.
This same mutation has also been shown to reduce the susceptibility of the B.1.1.7 (UK) and B.1.526 (New York) variants to the monoclonal neutralizing antibody bamlanivimab.
Furthermore, the B.1.427/B.1.429 variant that emerged in California contains an L452R mutation in the RBD that increases transmissibility by around 20% and has reduced neutralization by some EUA therapeutics.
The researchers say that although experimental studies into the potential threats of emerging variants exist, they have been limited to only a handful of known RBD mutations and to mAbs developed by the pharmaceutical companies Eli Lilly and Regeneron.
Furthermore, “there is no reliable measurement about whether a mutation will evade a vaccine because no one knows how many different antibodies will be created from the vaccination,” writes the team.
“Our approach helps close the gap”
“The current experimental capability lags behind the rapidly growing RBD mutations,” says Wei and colleagues. “Our approach helps close the gap.”
The team analyzed 261,348 genome sequences isolated from patients and identified 514 mutations in the spike RBD, 95 of which were high-frequency mutations.
Using machine learning and deep learning models validated with tens of thousands of experimental data points, the researchers performed a threat analysis to investigate the impacts of the mutations on 16 mAbs in clinical trials.
“The current experiments in the literature are limited to two EUA monoclonal antibody therapeutics from Regeneron and Eli Lilly,” says the team. “We extend our analysis to many other antibody therapeutic candidates that are in various stages of clinical trials, such as those from Celltrion and the Rockefeller University.”
What did the team find?
The study revealed that in addition to probable compromise by known emergent mutations, the Eli Lilly antibodies bamlanivimab and etesevimab are also likely to be compromised by the following high-frequency mutations: V483F/A, E484Q/V/A/G/D, F486L, F490L/V/S, Q493L, and S494P.
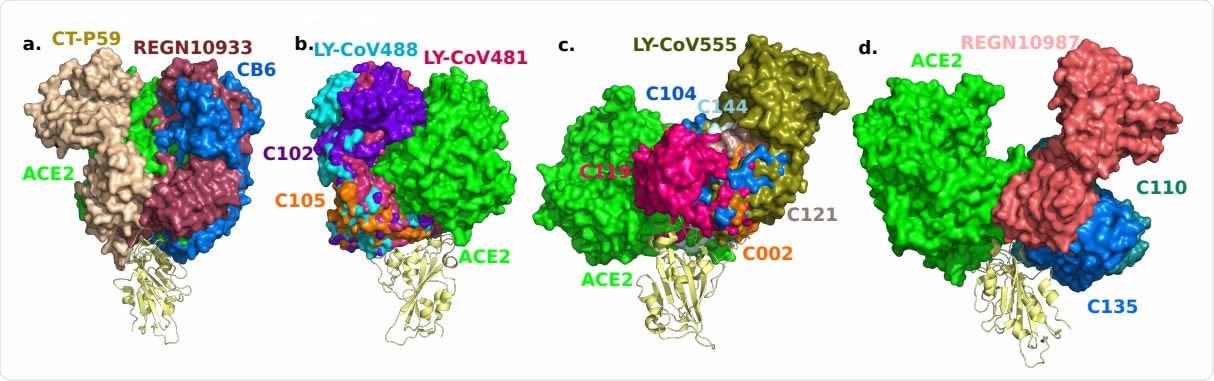
3D alignment of 16 antibodies and ACE2 on the S protein RBD. a. CT-P59 (7CM4), REGN10933 (6XDG), CB6 (7C01). b. LY-CoV488 (7KMH), LY-CoV481 (7KMI), C102 (7K8M), C105 (6XCM). c. LY-CoV555 (7KMG), C002 (7K8T), C104 (7K8U), C119 (7K8W), C121 (7K8X), C144 (7K90). d. REGN10987 (6XDG), C110 (7K8V), C135 (7K8Z).
Similarly, the Regeneron antibodies casirivimab and imdevimab are not only likely to be compromised by the known mutation K417T, but also the high-frequency mutations N439K, G446V, E484G, and F486L.
The study also predicted that while the Celltrion antibody regdanvimab will be compromised by variants P.1, B1.351, B.1.427, and B.1.526, it could also be rendered less effective by the high-frequency mutations L455F, E484A, F490L/S, and S494P/L.
Furthermore, the high-frequency mutations R346K/S could evade targeting by the Rockefeller University antibody C135.
In addition, the Rockefeller antibody C144 may also be compromised by the high-frequency mutations E484Q/A.
The team also identified more than 30 low-frequency RBD mutations with the potential to become vaccine or antibody escape variants in the future.
3D structure of human ACE2 (hACE2) and RBD. Color on the RBD structure indicates the BFE changes induced by mutations, where blue means binding strengthening and red means weakening.
The mutations pose “imminent threats”
“We unveil, for the first time, that high-frequency mutations R346K/S, N439K, G446V, L455F, V483F/A, E484Q/V/A/G/D, F486L, F490L/V/S, Q493L, and S494P/L might compromise some of the mAbs in clinical trials,” writes Wei and colleagues
“There are 95 fast-growing RBD mutations around the world that pose imminent threats to existing and future vaccines and antibody therapies,” they warn.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources