Researchers in the United States have predicted the binding epitopes of uncharacterized neutralizing antibodies that target the receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein.
The novel SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that continues to pose a threat to global public health and the economy worldwide.
The spike protein mediates the first stage of the SARS-CoV-2 infection process through binding of its RBD to the host cell receptor angiotensin-converting enzyme 2 (ACE2).
The study revealed whether RBD mutations enhanced the binding of ACE2 to improve viral infectivity or disrupted the binding of neutralizing antibodies (NAbs) to evade the host immune response.
The team found that of nine RBD variants tested, the N501Y, E484K, and K417T mutations, which are present in the Brazil P.1 and South Africa B.1.351 South Africa variants of concern (VoC), disrupted binding to 65% of the NAbs evaluated.
The researchers from the University of Alabama at Birmingham and the Texas Biomedical Research Institute in San Antonio also found that the L452R variant, which is found in the California B.1.427/B.1.429 VoC enhanced ACE2 binding while simultaneously disrupting certain classes of NAbs.
Furthermore, Mark Walter and colleagues say the analysis identified an antibody pair that bound to all RBD variants evaluated, representing an excellent opportunity for the treatment of both wildtype SARS-CoV-2 and VoC strains.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
More about the spike protein and NAbs
Human NAbs that bind to the SARS-CoV-2 spike RBD and block RBD–ACE2 interactions have been demonstrated as effective therapeutics. They are also a key part of the immune response elicited by vaccination.
On binding to host cells, the spike protein undergoes conformational changes that are essential for viral infectivity. The RBD switches from a “down” conformation, where the ACE2 binding site is buried within the protein, to an “up” conformation where this site is accessible for binding to the host cell surface.
“Presumably, the ‘down’ conformation of the RBD ‘hides’ the ACE2 binding site in the closed arrangement to, at least partially, protect the RBD from the host’s immune response,” says Walter and the team.
Since SARS-CoV-2 VoCs have been shown to reduce antibody-mediated neutralization, Walter and colleagues set out to further evaluate these single mutations at the molecular level.
What did the researchers do?
To date, the NAbs that target the SARS-CoV-2 spike RBD have been classified into four structural groups – C1 to C4 – based on their ability to bind to the “up” or “down” conformation and based on their overlap with the ACE2 binding site.
Using an RBD binding profile, the researchers classified NAb-RBD binding epitopes found in the protein databank as C1, C1D, C2, C3, or C4 and used this classification system to predict the binding epitopes of a series of uncharacterized NAbs isolated from a single convalescent patient.
Twenty unknown NAbs were assigned to a NAb-RBD complex and validated using RBD variant binding and epitope mapping studies.
Naturally occurring SARS-CoV-2 RBD sequence variation was also quantified to predict the binding sensitivities of the NAbs to RBD variants.
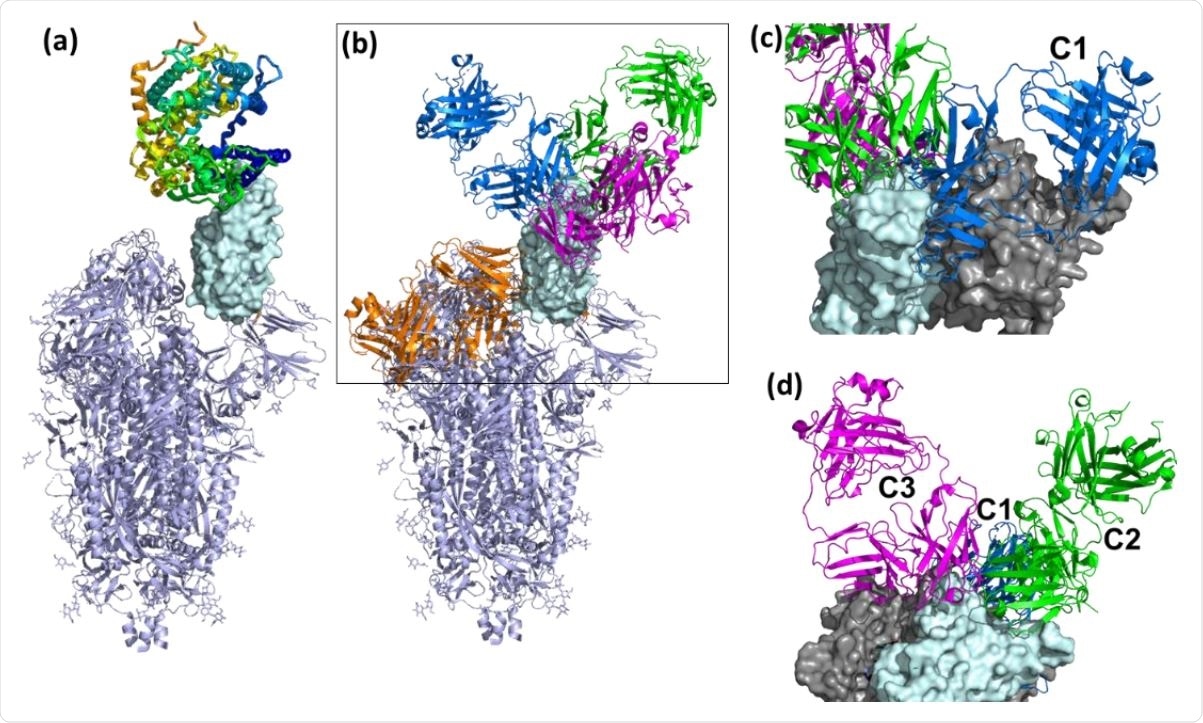
Overall view of ACE2 and NAbs binding to SARS-CoV-2 S protein. (a) Ribbon diagram of the SARS-CoV-2 S protein, with one RBD in the up conformation (cyan surface) interacting with ACE2. (b) General location of NAb binding epitopes, where an example C1 NAb is colored blue, C2 green, C3 magenta and C4 orange. The figure highlights that extensive conformational changes are required before C4 NAbs can bind to S. (c) View of two RBDs from an S trimer (cyan and grey surfaces) in the down conformation, to show that C1 NAbs cannot bind to their epitope while the RBD is in the down conformation, due to steric hindrance. (d) An alternative view of adjacent down RBDs in the S trimer showing C2 and C3 NAb epitopes are accessible while RBD is in the down conformation and that some NAbs can bind across an RBD-RBD trimer interface and interfere with RBD opening to bind ACE2.
What did the study find?
Of nine RBD variants tested, the K417T, E484K, and N501Y mutations present in the Brazilian P.1 and South African B.1.351 lineages disrupted the binding of 65% of the NAbs evaluated.
The study revealed that E484K was the most disruptive mutation, impacting the most significant proportion (30%) of NAbs tested. The N501Y and K417T mutations were only slightly less disruptive, impacting 25% of the NAbs.
In addition to disrupting NAb binding, E484K and N501Y exhibited binding affinities for ACE2 that were equivalent to that of a wild-type reference RBD variant.
However, the RBD variants N440Y and I345I also exhibited ACE2 binding affinity similar to wildtype RBD, suggesting that neither E484K nor N501Y increase infectivity by enhancing affinity for ACE2.
“If increased ACE2 affinity is required for increasing viral infectivity, other mutations in the SARS-CoV-2 spike must be responsible for enhanced ACE2 binding affinity,” says Walter and the team.
The K417T mutation also significantly disrupted NAb binding but exhibited a 2-fold reduction in ACE2 affinity, suggesting that this mutation is unlikely to confer a selective advantage through enhanced viral attachment.
Another RBD variant – L452R – which is associated with the California VoC B.1.427/B.1.429, was slightly less disruptive to Nab binding but showed a 2-fold increase in affinity for ACE2.
“This is consistent with the B.1.429 California VoC being able to evade NAb responses, as well as enhance ACE2 binding affinity, possibly providing advantages in immune evasion against NAbs, and increased cell infectivity via enhanced ACE2 binding affinity, ultimately leading to increased disease severity,” write the researchers.
One pair of NAbs could offer an “excellent” therapeutic option
The analysis also identified a pair of potent non-overlapping NAbs – 1213H7 and 1215D1 – that bound to all nine RBD variants evaluated.
Walter and colleagues say this represents an excellent potential therapeutic treatment for infection with wildtype SARS-CoV-2 and newly identified VoC.
Furthermore, “our study provides a strategy for characterizing NAb/RBD epitopes at a larger scale to further define NAb sensitivity to RBD variants in individuals recovered from natural infections and changes in the anti-RBD SARS-CoV-2 structural repertoire induced by vaccination,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources