Researchers in the United States and India have developed a high-volume, low-cost platform for manufacturing vaccines to protect against different variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The new manufacturing process uses a genetically engineered strain of the microbe Komagataella phaffii (Pichia pastoris) to produce the spike RBD. The use of this microbial host eliminates the need for methanol-induction of the recombinant RBD gene, which can present challenges in large-scale facilities.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team from the Massachusetts Institute of Technology and the Serum Institute of India Pvt. Ltd. in Pune says that this methanol-free production increased secreted titers of the RBD protein by more than five times.
“Strains engineered for use without methanol and increased productivity could facilitate manufacturing of RBD and other antigens for vaccine candidates at large volumes and low costs to enable accessible and affordable vaccines for global use,” writes Christopher Love and colleagues.
“This engineered strain is now used to produce an RBD-based vaccine antigen that is currently in clinical trials and could be used to produce other variants of RBD as needed for future vaccines,” adds the team.
A pre-print version of the research paper is available on the bioRxiv* platform, while the article undergoes peer review.
New manufacturing platforms will be needed to keep up with demand
As new variants of SARS-CoV-2 continue to emerge, prevention of COVID-19 on a global scale will require the ongoing development of high-volume, low-cost vaccine manufacturing platforms to keep up with continued demand.
Vaccine candidates based on recombinant protein subunits can be manufactured at low cost in existing large-scale production facilities that use microbial hosts such as K. phaffii.
“The 201 amino acid RBD is an especially promising antigen for accessible vaccines because it can be manufactured at low cost and high volumes in microbial hosts,” writes Love and the team.
The methylotrophic yeast strain K.phaffii is routinely used to produce large volumes of therapeutic proteins. A key advantage of this production host is the tightly regulated methanol-inducible promoter – PAOX1 – that is used for the expression of the recombinant gene.
“This promoter enables outgrowth to high cell densities with inexpensive feedstock like glycerol before induction of the recombinant gene with methanol feed,” say the researchers.
However, the use of methanol presents challenges in large-scale facilities, including high heat generation during fermentation and flammability concerns regarding storage.
These challenges mean that such facilities require specific designs or modifications that enable the use of methanol, potentially limiting the number that can produce vaccine components such as RBD antigens in K. phaffii.
What did the researchers do?
To assess the feasibility of methanol-free production, Levy and colleagues cultivated a genetically engineered strain of K. phaffii expressing the RBD (Wuhan-Hu-1 sequence) regulated under the native AOX1 promoter.
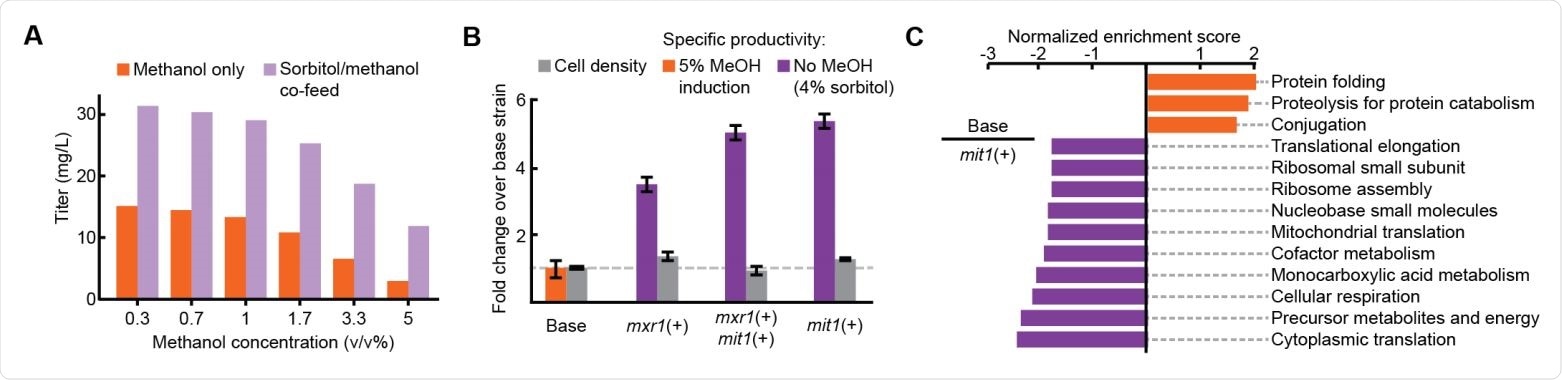
Improved productivity and decreased stress in methanol-free RBD expression (A) Titer of RBD secretion from the base strain in 3 mL plate culture. (B) Performance of three engineered strains in 3 mL plate culture. (C) Enriched gene sets between the base strain (orange) and the mit1+ strain (purple).
The team induced expression of the recombinant gene with different amounts of methanol. Interestingly, this resulted in higher titers of RBD, the more the methanol concentration was reduced.
Next, the researchers induced protein production with a mixture of methanol and sorbitol – a supplementary carbon source that does not repress PAOX1 expression.
This further increased the RBD titer, which led the team to hypothesize that efficient secretion of the RBD could be achieved without using any methanol.
Levy and colleagues say that several studies have shown that constitutive overexpression of activating transcription factors can lead to consistent activation of PAOX1, without the use of methanol.
They, therefore, integrated additional copies of the transcription factors mit1 and mxr1 into the K. phaffii genome and cultivated these strains for protein production using only sorbitol.
RBD productivity was increased more than 5-fold
This approach increased RBD productivity more than 3-fold across all strains and more than 5-fold in a strain that contained just one extra copy of the transcription factor mit1.
Next, the team generated a mit1+ strain that expressed RBD with the fusion of SpyTag and transferred this RBD-SpyTag encoding strain to a facility for good manufacturing practices (GMP) production in a 1,200 L fed-batch process.
“Removal of methanol from the production process enabled scale up to a 1,200 L pre-existing production facility,” write the researchers.
At this facility, the strain produced 21mg of purified, clinical quality RBD-SpyTag per liter of fermentation or more than 1 million doses from a single reactor batch.
The modified mit1+ strain also increased productivity of other variants
Next, Levy and colleagues assessed whether this modified mit1+ strain could also increase the production of sequence variants for other circulating strains of SARS-CoV-2. They generated strains expressing the B.1.1.7 (UK) and B.1.351 (Brazil) RBD variants and evaluated their specific productivities.
This revealed that for all RBD variants, the mit1+ engineered strain improved productivity in the absence of methanol.
“This result demonstrates that the engineered mit1+ strain could facilitate new cell lines for manufacturing other RBD variants without methanol for seasonal vaccine boosters or next-generation vaccine candidates for emerging variants,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Love C, et al. Scalable, methanol-free manufacturing of the SARS-CoV-2 receptor binding domain in engineered Komagataella phaffii. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.04.15.440035, https://www.biorxiv.org/content/10.1101/2021.04.15.440035v1
- Peer reviewed and published scientific report.
Dalvie, Neil C., Andrew M. Biedermann, Sergio A. Rodriguez‐Aponte, Christopher A. Naranjo, Harish D. Rao, Meghraj P. Rajurkar, Rakesh R. Lothe, et al. 2021. “Scalable, Methanol‐Free Manufacturing of the SARS‐CoV‐2 Receptor‐Binding Domain in Engineered Komagataella Phaffii.” Biotechnology and Bioengineering 119 (2): 657–62. https://doi.org/10.1002/bit.27979. https://onlinelibrary.wiley.com/doi/10.1002/bit.27979.