Human-to-human transmission of SARS-CoV-2 triggered an ongoing pandemic of the coronavirus disease 2019 (COVID-19) with unprecedented impact on health and economic systems worldwide. However, there is still no specific treatment for patients that develop a severe form of the illness.
And while vaccines definitely provide a promising path of disease prevention, as well as substantially curbing the infection rates, there is still a dire need to develop and implement specific therapeutic agents in order to reduce the most severe consequences from infection and ensuing patient mortality.
One possibility is microRNAs (miRNAs), which are non-coding RNAs involved with the regulation of post-transcriptional gene expression that may impact entire pathways related to viruses and diseases they can cause. Consequently, advances in RNA chemistry and delivery possibilities opened the door for the first miRNA-based agents to enter into clinical trials.
More recently, several studies have appraised differential expression of miRNAs in COVID-19 patients and suggested their use as biomarkers or therapeutics. Lung biopsies in nine COVID-19 patients demonstrated that miR-26a, miR-29b, and miR-34a actually correlate with inflammatory biomarkers and endothelial dysfunction.
In this new study, currently available on the bioRxiv* preprint server, a large group of researchers (led by Dr. J. Tyson McDonald from the Georgetown University School of Medicine) started a quest for miRNAs that may directly regulate and drive a COVID-19 response.
From –omics approaches to animal models
In order to pinpoint miRNAs that may be directly implicated in driving COVID-19 severity in the host, the researchers initially examined publicly available bronchial alveolar lavage fluid RNA-sequencing data from thirteen individuals.
Likewise, to ascertain whether miR-2392 might be able to drive the observed COVID-19 symptoms and complications in the infected host, they have analyzed the conservation of specific human miR-2392 across species and its integration into the SARS-CoV-2 genome.
Further pathway analysis was carried out with targets and pathways for miR-2392 in order to determine its impact on the host when upregulated. One of the most important steps was detailed proteomic and transcriptomic analysis on miR-2392 targets on blood from COVID-19 patients using the COVIDome dataset.
To reveal the presence of circulating miR-2392 in infected individuals, the scientists have also quantified the amount of miR-2392 by droplet digital PCR in the obtained sera, nasopharyngeal swab samples, and urine. Finally, in vitro human and in vivo hamster models were used to test therapeutic inhibitors targeting this microRNA.
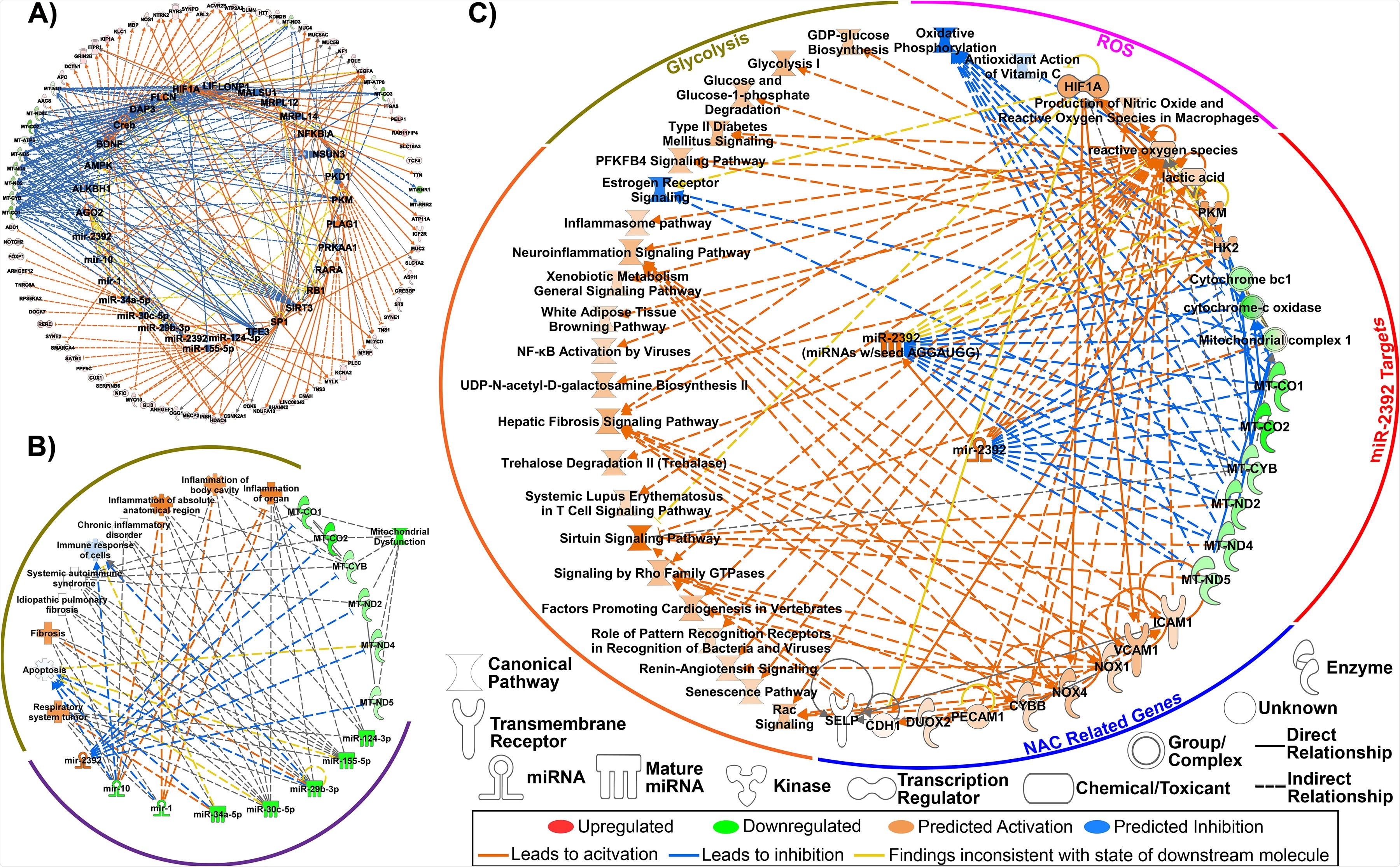
Key miRNA signature as predicted from Bronchial Alveolar Lavage Fluid (BALF) RNA-seq data in patients with COVID-19. A) Predicted upstream regulators determined through Ingenuity Pathway Analysis (IPA) consistent with the transcriptional response from differentially expressed genes (FDR<0.05; outer ring). Eight miRNAs were among the key regulators in response to COVID-19 (inner ring). B) Major biological responses resulting from dysregulation of this eight miRNA signature drive immune- and inflammatory719 related pathways as well as mitochondrial dysfunction determined through IPA. C) Pathway regulation by miR-2392 from BALF RNA-seq data determined through IPA.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
miR-2392 as an effective biomarker of COVID-19
In short, this study uncovered eight new miRNA signatures in patients with COVID-19 viral loads (compared to healthy individuals), as predicted from RNA sequencing data. More specifically, the expression of seven miRNAs was decreased, but one single was significantly increased – and that was the aforementioned miR-2392.
Additional examination unveiled miR-2392 as a key miRNA involved in COVID-19 progression. More specifically, the study has clearly shown how miR-2392 can drive downstream suppression of mitochondrial activity while at the same time increasing inflammation, glycolysis (i.e., breakdown of glucose for energy extraction), and hypoxia (i.e., inadequate oxygen supply).
Moreover, the upregulation of miR-2392 was observed concomitantly with symptoms linked to COVID-19 infection in the host. Also, the researchers have found that miR-2392 was circulating in COVID-19 infected patients and increased as a function of viral load.
The added value of miRNA
These results point towards the conclusion that miR-2392 may be used as an effective biomarker of COVID-19. But this study went one step further, as researchers have actually developed a miR-2392 inhibitor and provided evidence that its use reduces SARS-COV-2 viability in used cell cultures and animal models.
“With further development, this miR-2392 inhibitor may represent an effective antiviral therapeutic towards inhibiting the virus and limiting a negative host response from COVID-19”, explain study authors in this bioRxiv paper. “This key miRNA signature was involved in major cellular and molecular mechanisms that drives the viral-host response”, they conclude.
As the testing of drug-targeted and antibody-based therapies are currently ongoing, the added value of miRNAs represents a new wave of treatment compounds that have previously shown endogenous activity to modify the course of viral infections.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
McDonald, T.J. et al. (2021). The Great Deceiver: miR-2392’s Hidden Role in Driving SARS-CoV-2 Infection. bioRxiv. https://doi.org/10.1101/2021.04.23.441024, https://www.biorxiv.org/content/10.1101/2021.04.23.441024v2
- Peer reviewed and published scientific report.
McDonald, J. Tyson, Francisco J. Enguita, Deanne Taylor, Robert J. Griffin, Waldemar Priebe, Mark R. Emmett, Mohammad M. Sajadi, et al. 2021. “Role of MiR-2392 in Driving SARS-CoV-2 Infection.” Cell Reports 37 (3). https://doi.org/10.1016/j.celrep.2021.109839. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)01303-6.