Whole-genome sequencing studies of newly emerging viral variants are particularly important for identifying mutations that emerge under positive selection and play vital roles in viral evolution. In the later phase of the coronavirus disease 2019 (COVID-19) pandemic, several novel SARS-CoV-2 variants with multiple spike protein mutations have emerged. Because of significantly increased infectivity and immune escape ability, some of these variants have become the predominantly circulating variants worldwide. Therefore, to effectively control the trajectory of COVID-19 pandemic, continuous genomic surveillance of SARS-CoV-2 sequences both at the national- and global level are of prime importance.
In the current study, scientists have reported the emergence of a novel SARS-CoV-2 variant containing signature mutations in the membrane protein-encoding gene.
Study design
The scientists conducted reverse transcription-polymerase chain reaction (RT-PCR) for whole-genome sequencing of a total of 2,900 SARS-CoV-2-positive patient samples. They subsequently compared these sequences with full-length SARS-CoV-2 sequences downloaded from the GISAID and NCBI databases for viral variant analysis.
Furthermore, they performed phylogenetic analysis and SARS-CoV-2 membrane protein structural predictions using a multiple sequence alignment program and a specialized service for structural analysis of missense variants.
Important observations
By analyzing the missense mutation carrying genome and synonymous mutation carrying genome, the scientists estimated that missense mutations should occur 2.7 times, 3.1 times, 3.8 times more frequently than the synonymous mutations in the envelope protein-coding gene, membrane protein-coding gene, and ORF6-coding gene, respectively. By comparing missense mutation carrying genomes with synonymous mutation carrying genomes, they noticed that the membrane gene is highly conserved and possibly under robust purifying selection.
By estimating the membrane gene missense mutation carrying viral genomes, they identified only 4 missense mutations in viral genomes collected from the USA in February 2021. With further analysis, they observed a gradual increase in missense membrane gene mutations in viral genomes in the USA and worldwide.
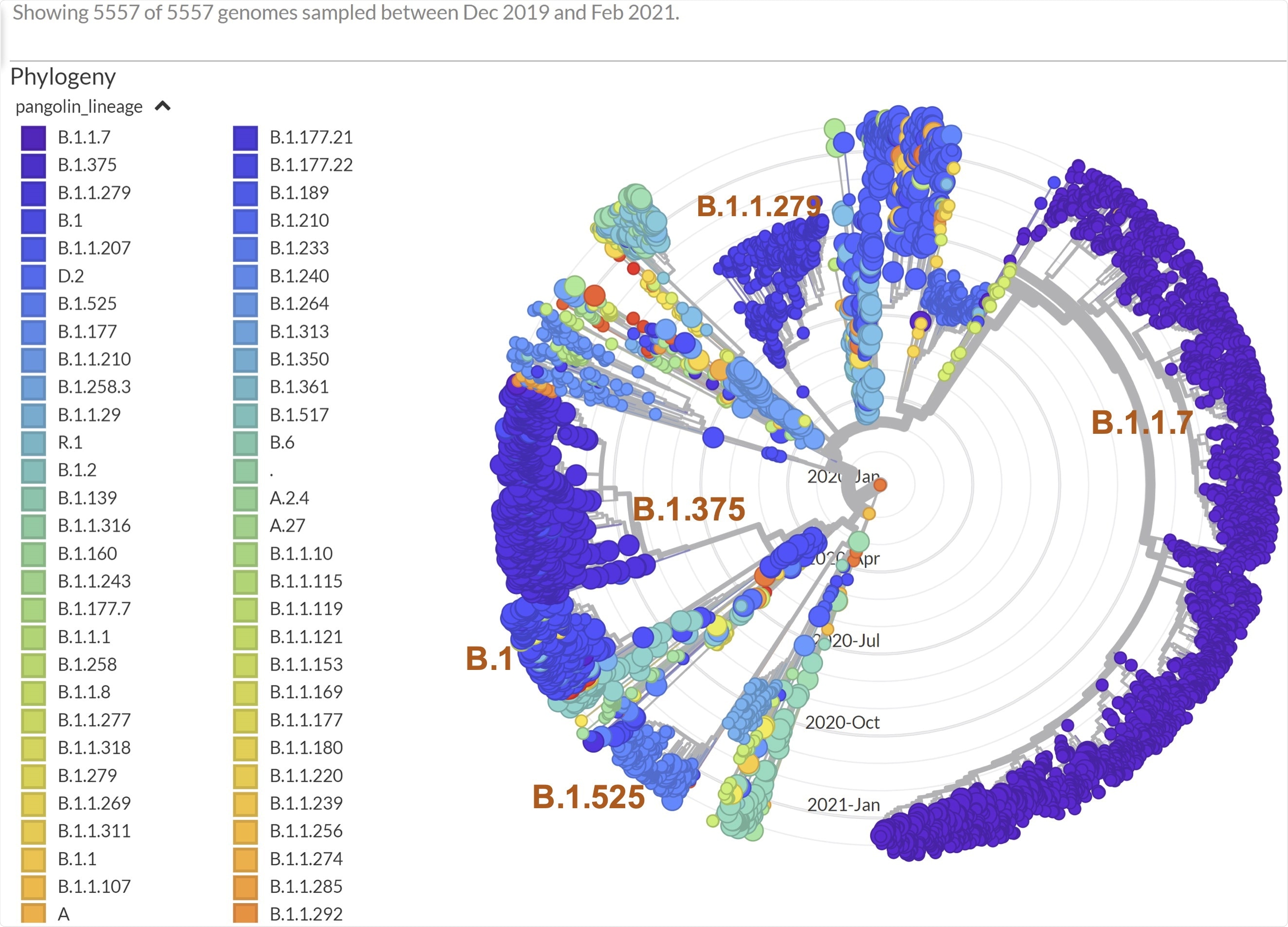
Phylogenetic analysis of viral genomes carrying missense M mutations, colored by lineage background.
By analyzing the frequency of increase for these mutations, they observed that M:I48V mutation is highly specific to the USA and that most of the viral isolates carrying this mutation belong to the B.1.375 lineage. However, currently, the frequency of this mutation dropped significantly in the USA.
In contrast to the M:I48V mutation, the frequency of M:I82T mutation was found to increase by 116-fold between October 2020 and February 2021 in the USA. The scientists confirmed that the viral isolates carrying this mutation predominantly belong to the B.1 and B.1.525 lineages and that about 99% of the B.1.525 lineage currently carry this mutation.
With further analysis, the scientists noticed that the largest M:I82T carrying clade is part of a young M:I82T sub-B.1 lineage and that about 90% of the isolates in this clade contain 10 other missense mutations. They mentioned that the M:I82T clade is significantly different from other B.1 lineage clades, and thus, a separate designation is required for this clade. Interestingly, they found that the second-largest M:I82T carrying clade is currently circulating in Europe and Africa and co-segregating with the E484K Spike protein mutation. Because of the absence of E484K Spike mutation in the USA M:I82T clade, the scientists suggest that M:I82T mutation may have implications in viral evolution independent of E484K Spike mutation.
The scientists identified a number of other membrane gene mutations that showed a recent surge in frequency worldwide. Focusing on these mutations, they collected more than 5,500 sequences carrying any of these mutations for phylogenetic analysis. Their analysis revealed that many of these sequences belong to the B.1.1.7 lineage, which also carries a synonymous membrane gene mutation.
Furthermore, the scientists selected four membrane gene mutations (M:A2S, M:F28L, M:I82T, and M:V70L) and analyzed their presence in viral isolates collected from the study patients. Their analysis revealed that the mean age of patients carrying each of these mutations was 37 – 39 years, whereas the mean age of patients without these mutations was 43 years.
Study significance
The study highlights the rapid emergence of the B.1.I82T clade with SARS-CoV-2 membrane gene mutations in the USA and worldwide. Overall, the findings suggest that viral variants containing membrane gene mutations exhibit higher transmissibility among younger populations and that mutations emerging in the viral membrane protein-encoding gene play crucial roles in increasing viral fitness probably by controlling glucose uptake during viral replication.