Most of the major life-threatening symptoms of COVID-19 disease are associated with the host's generation of an exacerbated pro-inflammatory immune response. Many studies have correlated the intensity of the immune response with the severity of disease.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters cells via the angiotensin-converting enzyme 2 (ACE2) receptor, commonly found on the surface of cells in the lungs, though also with a significant presence in the gastrointestinal tract, where there is evidence for replication of the virus due to the strong presence of viral RNA in feces. Interestingly, some reports have suggested that the cellular response of cells in either the lungs or gut is distinct upon SARS-CoV-2 infection. The former demonstrates a curtailed interferon response, while intestinal epithelial cells seemingly increase production.
In a paper recently published in the journal Molecular Systems Biology by Alexandro et al. (April 27th, 2021) single-cell RNA sequencing is applied to the human ileum- (the final section of the small intestine) and colon-derived organoids infected with SARS-CoV-2 in an effort to better understand virus tropism towards specific cell types, with surprising findings regarding the most commonly infected types of cell.
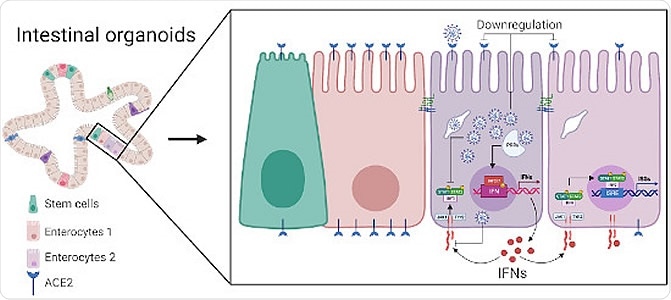
Single-cell sequencing and multiplex single‐molecule RNA FISH analyses on SARS‐CoV‐2 infected human intestinal organoids characterize the tropism of SARS‐CoV‐2 and identify strategies developed by the virus to interfere with the host intrinsic innate immune response.
SARS-CoV-2 gut cell specificity
Following the generation and infection of the human ileum and colon organoids, the group noted that only around 10% of the cells would become infected, even when higher viral loads were applied. These discrete populations were observed to be infected from four hours post-application onwards, steadily increasing over the course of the 24-hour experiment, and exhibited upregulated interferon λ-2 and -3, while interferon λ-1 and β-1 were unaltered from baseline levels. Initially, it was thought that around 75% of cells were infected at 24 hours by RNA sequencing. However, parallel immunofluorescence staining revealed that most of these cells were not infected, viral particles or free viral RNA instead being bound to the cell surface or floating nearby. Correcting for these background levels of bound RNA allowed the group to ascertain that only 4.5% and 5.3% of colon and ileum cells were truly infected, respectively.
The ileum and colon organoids were composed of eight and nine major populations of cells, respectively. Both were composed of stem cells, transient amplifying cells, enterocytes, and enteroendocrine cells, with the ileum bearing a more significant proportion of goblet cells. Importantly, these ratios and the clustering of the cells were found to be consistent with biopsies of these organs collected from COVID-19 patients, allowing the cellular tropism of SARS-CoV-2 to be studied under representative conditions.
Immature enterocytes 2 were the most consistently infected cells in both organoids, with transient amplifying cells being secondary targets, infected only at 24 hours in the ileum organoid. Interestingly, amongst all of the cells included in the organoids, immature enterocytes 2 do not display the greatest levels of the ACE2 receptor. Immature enterocytes 1 display the receptor significantly more commonly and did not appear to be particularly susceptible to SARS-CoV-2 infection. TMPRSS2 is a cellular protease that is essential to SARS-CoV-2 envelope fusion with the host membrane. This protein was noted to be in abundance in immature enterocyte 2 cells, the concentration correlating with SARS-CoV-2 genome copy number in the infected cells. The group speculate that this protein may play a more significant role than was previously suspected, perhaps surpassing the ACE2 receptor in importance.
Further, it was observed that those cells infected with SARS-CoV-2 underwent reduced ACE2 receptor expression, rather than increased as seen in lung-derived cells, and upregulated tumor necrosis factor and NFκB cytokine production, inducing interferon release from bystander cells. The colon organoids exhibited an initial increase in ACE2 expression at 12 hours, and an overall lesser expression at 24 hours, while the ileum-derived organoids showed gradually lesser expression over the 24 hours. The group suggests that ileum organoids are more immunoresponsive than those derived from the colon, producing significantly more interferon-stimulated gene products. This is supported by other studies of bacterial and viral challenges to the gastrointestinal tract. As observed, only bystander cells generate interferon upon SARS-CoV-2 infection. Thus the virus appears to have developed the capacity to suppress interferon-mediated signaling in gut cells, as has also been observed in the lungs.
In conclusion, the authors suggest that SARS-CoV-2 is able to lessen the immune response of infected cells in the gut, allowing greater pathogenicity, while responsive bystander cells act as a pro-inflammatory reservoir that may contribute towards the more severe symptoms of COVID-19.
Journal reference:
- Single‐cell analyses reveal SARS‐CoV‐2 interference with intrinsic immune response in the human gut, Sergio Triana, Camila Metz‐Zumaran, Carlos Ramirez, Carmon Kee, Patricio Doldan, Mohammed Shahraz, Daniel Schraivogel, Andreas R Gschwind, Ashwini K Sharma, Lars M Steinmetz, Carl Herrmann, Theodore Alexandrov, Steeve Boulant, Megan L Stanifer, Mol Syst Biol. (2021) 17: e10232, https://doi.org/10.15252/msb.202110232, https://www.embopress.org/doi/full/10.15252/msb.202110232