The most severe symptoms of COVID-19 are associated with the dysregulation of immune functions in the respiratory tract, in particular, leading to excessive inflammation as the result of a cytokine storm. Upon infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) respiratory epithelial cells or alveolar macrophages respond by producing interferon and various pro-inflammatory cytokines, such as interleukin-6 and tumor necrosis factor-α.
A variety of immune cells then flood the tissue, including neutrophils, dendritic cells, and monocytes, which express lymphocyte antigen 6 (Ly6C) variably amongst the population. Monocytes presenting high levels of Ly6C tend to be pro-inflammatory, and upon infection with SARS-CoV-2, Ly6C-high monocytes infiltrate the lungs in a CC chemokine receptor type 2 (CCR2) dependent manner.
In a paper recently uploaded to the preprint server bioRxiv* by Vanderheiden et al. (May 4th, 2021) the contribution of monocytes in promoting protective immunity from SARS-CoV-2 is investigated in a murine model, suggesting that CCR2 plays the role of an inflammation modulator in early infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
CCR2 and inflammatory markers
The group began by developing a mouse-adapted SARS-CoV-2 strain, engineering mutations to the genome that have previously been demonstrated to enhance virulence in mice, including the N501Y mutation to the spike protein observed in the B.1.351 variant. Subsequently, the virus was passaged 20 times in the lungs of mice, and examination of the resulting genome revealed three newly acquired mutations by this process, two of which were within the spike protein (K417N and H655Y). The K417N mutation is also present on the B.1.351 variant, leading the group to attempt to infect mice with this strain unaltered in the lab. It was noted that this strain was able to replicate in C57Bl/6 mice to high viral load, producing similar levels of cytokines as the engineered mouse-adapted strain.
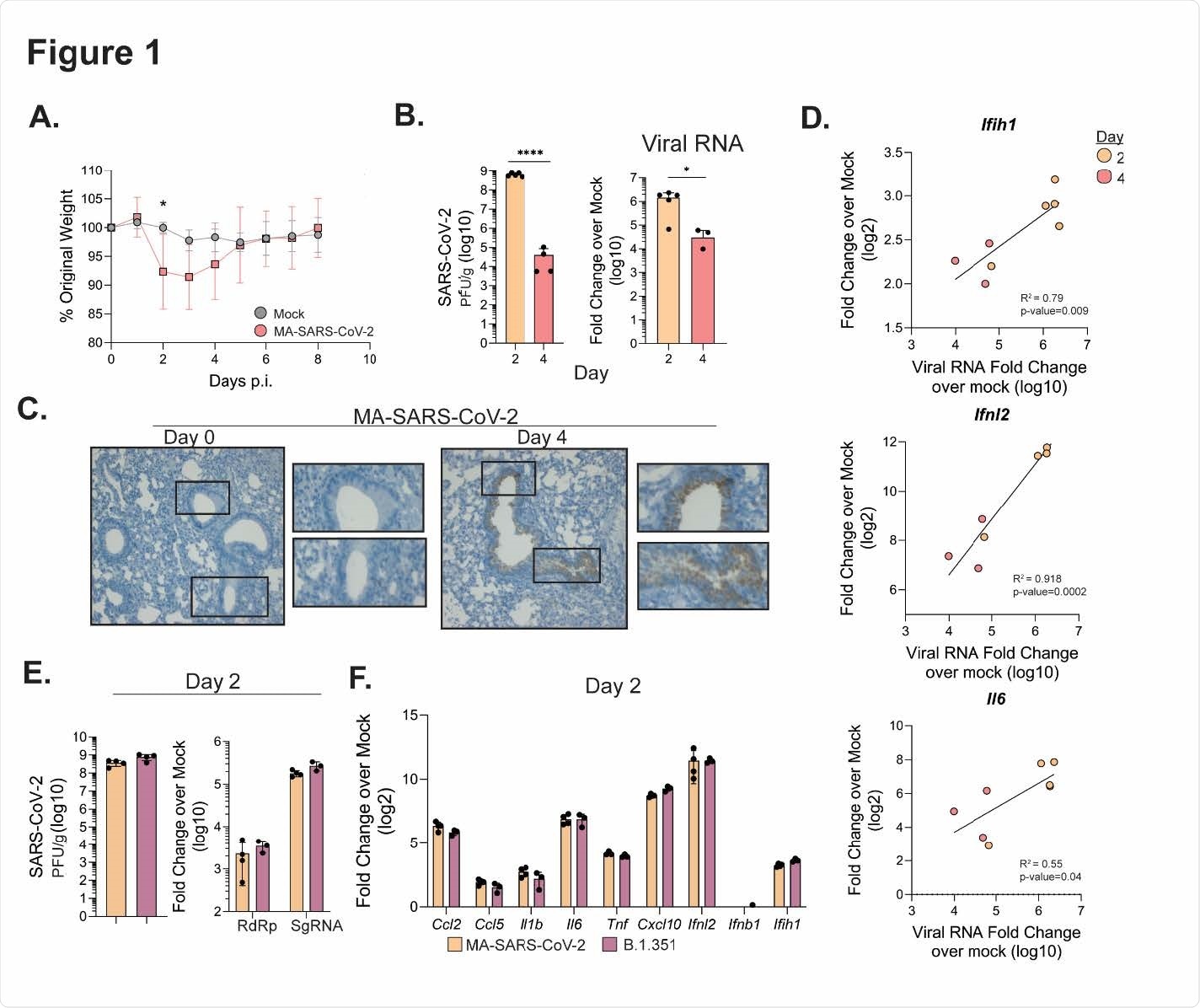
Figure 1. MA-SARS-CoV-2 and B.1.351 replicate in the respiratory tract. C57Bl/6J mice were infected intranasally with 5 x 105 PFU of MA-SARS-CoV-2 or an equal volume of PBS for mock mice. A) Percentage of initial weight for mock and MA-SARS-CoV-2 infected mice over 8 days. B) Quantification of MA-SARS-CoV-2 titers from lung tissue at the indicated day post infection as measured by plaque assay (left) or qRT-PCR (right). CT values are represented as relative fold change over mock (log10). C) In situ hybridization was performed using a probe for MA-SARS CoV-2 spike protein RNA. Representative images of lung slices from mock or day 4 p.i. D) Fold change over mock for the indicated gene was plotted against the corresponding MA-SARS-CoV-2 viral RNA for each sample, and linear regression was used to determine correlation. E) Quantification of viral titers from lung tissue by plaque assay at day 2 p.i. from mice infected with MA-SARS-CoV-2 or human variant B.1.351 (5 x 105 PFU/mouse). On the right, quantification of RNA-dependent RNA-polymerase (RdRp) or subgenomic RNA (SgRNA) fold change over mock. F) Gene expression measured via qRT-PCR for the indicated gene from lungs infected with MA SARS-CoV-2 or B.1.351 at 2 days p.i. Results are representative of 2 independent experiments with 5 mice per group. Statistical significance was determined using unpaired student’s t-tests or linear regression. * p<0.05, ****p<0.0001.
As observed in humans, there was a correlation between inflammatory cytokine levels and COVID-19 disease severity. The lungs exhibited an enhanced concentration of pyrogens, chemoattractants for monocytes, and T-cells interferon-stimulated genes, alarmins, and matrix metalloproteinases.
A significant shift in the composition of macrophages in the lungs was noted by the group in the days following infection, with a decrease in alveolar macrophages and an increase in activated transitional macrophages.
The total number of dendritic cells circulating remained constant while increasing ten-fold in the lungs, and other inflammation markers were observed to similarly increase in local concentration.
CCR2 knockdown mice experienced a more significant viral load in the lungs, and also bore higher neutrophil and inflammatory cytokine levels, which could be the direct result of knockdown or as a consequence of greater viral load.
Fewer monocytes or monocyte-derived macrophages entered the lungs of these knockdown mice, however, and there was also an observed ten-fold drop in transitional macrophage numbers compared to wildtype mice, suggesting that the CCR2 receptor plays a specific role in promoting recruitment and differentiation of these cells.
Interestingly, while virus RNA was found to be localized to the lining of the airways in wild-type mice, CCR2 knockdown mice exhibited much greater virus infiltration of the lungs, RNA being found much further into the interstitial spaces.
Lower viral burden and lesser weight loss was observed in mice with CCR2 while infected with the mouse-adapted SARS-CoV-2, and the group suggests that the ability of monocyte-derived cells to enter the lung parenchyma via CCR2 is crucial to limiting both viral load and cytokine overproduction in the early stages of infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
CCR2-dependent monocyte-derived cells restrict SARS-CoV-2 infection, Abigail Vanderheiden, Jeronay Thomas, Allison L Soung, Meredith E Davis-Gardner, Katharine Floyd, Fengzhi Jin, David A Cowan, Kathryn Pellegrini, Adrian Creanga, Amarendra Pegu, Alexandrine Derrien-Colemyn, Pei-Yong Shi, Arash Grakoui, Robyn S Klein, Steven E Bosinger, Jacob E Kohlmeier, Vineet D Menachery, Mehul S Suthar, bioRxiv, 2021.05.03.442538; doi: https://doi.org/10.1101/2021.05.03.442538, https://www.biorxiv.org/content/10.1101/2021.05.03.442538v1
- Peer reviewed and published scientific report.
Vanderheiden, Abigail, Jeronay Thomas, Allison L. Soung, Meredith E. Davis-Gardner, Katharine Floyd, Fengzhi Jin, David A. Cowan, et al. 2021. “CCR2 Signaling Restricts SARS-CoV-2 Infection.” Edited by Stacey Schultz-Cherry. MBio, November. https://doi.org/10.1128/mbio.02749-21. https://journals.asm.org/doi/10.1128/mBio.02749-21.